Figure 7.
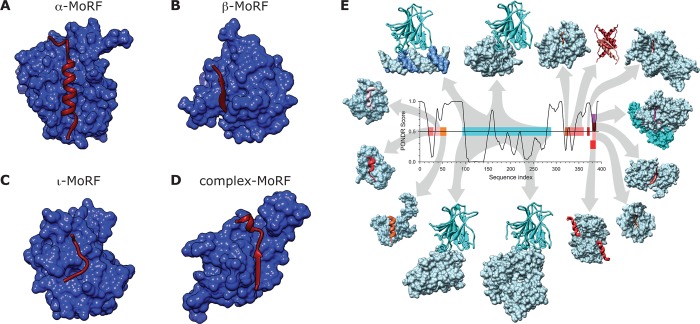
Classification of molecular recognition features (MoRFs) based on the secondary structure of the bound state. MoRFs (red ribbons) undergo disorder-to-order transition upon binding their partners (blue surfaces). (A) α-MoRF. BH3 domain of BAD (MoRF) bound to bcl-xl (partner) (PDB ID: 1G5J). (B) β-MoRF. Inhibitor of apoptosis protein DIAP1 (partner) bound to N-terminus of cell death protein GRIM (MoRF) (PDB ID: 1JD5). (C) ι-MoRF. AP-2 (partner) bound to the recognition motif of amphiphysin (MoRF) (PDB ID: 1KY7). (D) Complex-MoRF. Phosphotyrosine-binding domain (PTB) of the X11 protein (partner) bound to amyloid β A4 protein (MoRF) (PDB ID: 1X11). Note that the PTB domain of X11 actually binds unphosphorylated peptides and is a PTB by sequence similarity. Panels A–D reprinted with permission from ref (122). Copyright 2007 American Chemical Society. (E) Promiscuity of disorder-controlled interactions illustrated by the p53 interaction network. A structure versus disorder prediction on the p53 amino acid sequence is shown in the center of the figure (up = disorder, down = order) along with the structures of various regions of p53 bound to 14 different partners. The predictions for a central region of structure, and the disordered amino and carbonyl termini have been confirmed experimentally for p53. The various regions of p53 are color coded to show their structures in the complex and to map the binding segments to the amino acid sequence. Starting with the p53–DNA complex (top, left, magenta protein, blue DNA), and moving in a clockwise direction, the Protein Data Bank147 IDs and partner names are given as follows for the 14 complexes: (1tsr – DNA), (1gzh – 53BP1), (1q2d – gcn5), (3sak – p53 (tetramerization domain)), (1xqh – set9), (1h26 – cyclin A), (1ma3 – sirtuin), (1jsp – CBP bromo domain), (1dt7 – s100ββ), (2h1l – sv40 Large T antigen), (1ycs – 53BP2), (2gs0 – PH), (1ycr – MDM2), and (2b3g – RPA70). Reprinted with permission from ref (40). Copyright 2010 Elsevier.