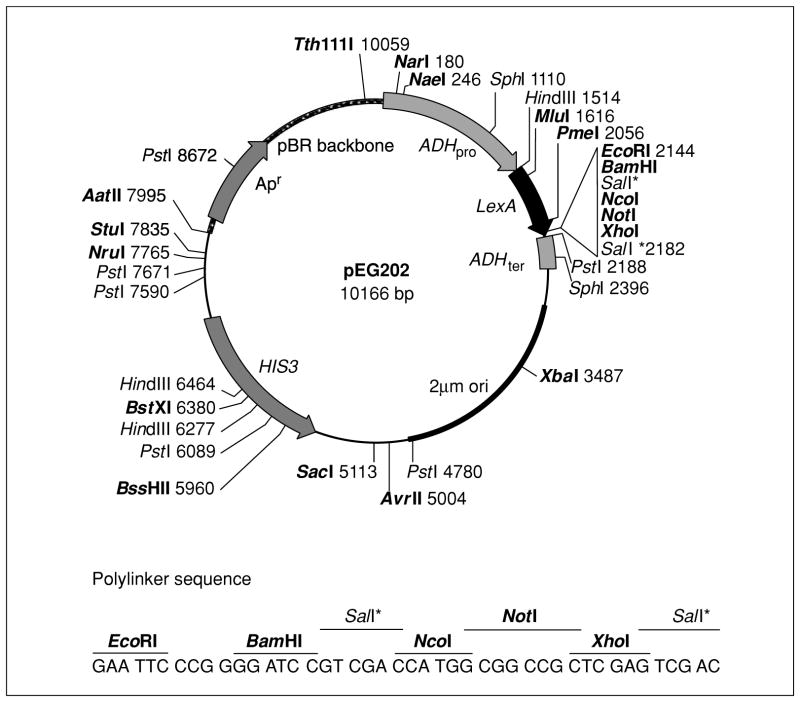
Figure 17.3.3.
LexA fusion plasmid pEG202. The strong constitutive ADH promoter is used to express bait proteins as fusions to the DNA-binding protein LexA. Restriction sites shown in this map are based on pEG202 sequence data and include selected sites suitable for diagnostic restriction endonuclease digests. A number of restriction sites are available for insertion of coding sequences to produce protein fusions with LexA; the polylinker sequence and reading frame relative to LexA are shown below the map with unique sites marked in bold type. The sequence 5′-CGT CAG CAG AGC TTC ACC ATT G-3′ can be used to design a primer to confirm correct reading frame for LexA fusions. Plasmids contain the HIS3 selectable marker and the 2-μm origin of replication to allow propagation in yeast, and the Apr antibiotic resistance gene and the pBR origin of replication to allow propagation in E. coli. In the plasmids pMW101 and pMW103, the ampicillin resistance gene (Apr) has been replaced with the chloramphenicol resistance gene (Cmr) and the kanamycin resistance gene (Kmr), respectively (see Table 17.3.1 for details).