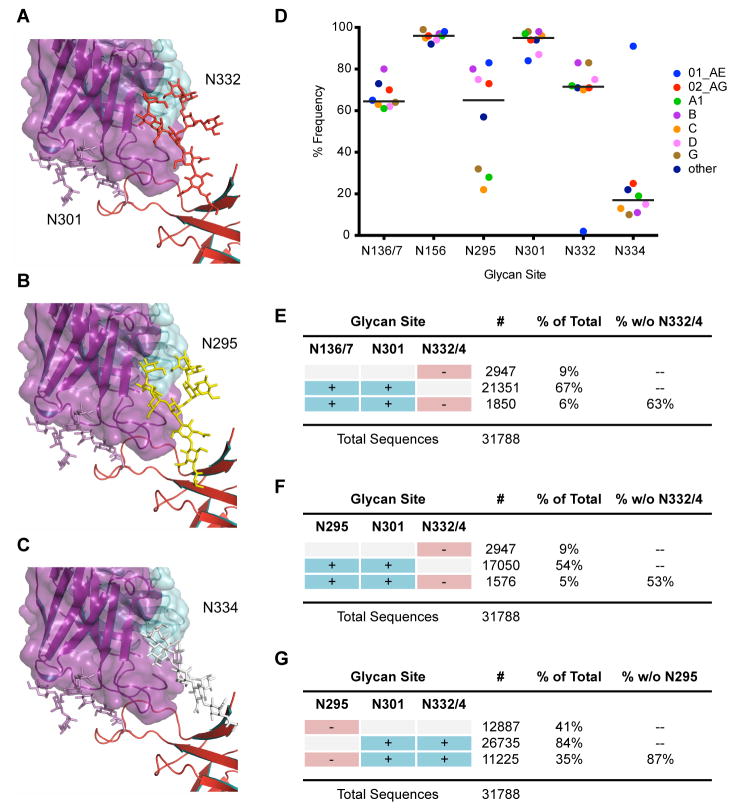
Figure 7. Structural modeling of high-mannose patch glycan sites and calculations of glycan site frequencies.
Glycan models to rationalize PGT128 promiscuity. (A) Close-up view of PGT128 binding to eODmV3 including the glycans at position N301 and N332, as defined by the crystal structure of eODmV3+PGT128. (B) Model for an N-linked Man8GlcNAc2 glycan at position N295 on eODmV3. (C) Model for an N-linked Man8GlcNAc2 glycan at position N334 on eODmV3. Coloring is as follows: PGT128, magenta; eODmV3, red; N301 glycan, magenta; N332 glycan, red; N295 glycan, yellow; N334 glycan, white. Cross-clade frequency of glycans at different positions on Env. (D) Frequency of critical N-linked glycan sites in 31,788 viruses in the Los Alamos database. A glycan site was determined as being present accordingly to the Nx(T or S) motif, where x is not a proline. Only clades for which > 100 sequences were available are included in the graph. The line represents the mean of the frequency for each glycan site. (E–F) Frequency of combinations of the N-linked glycan sites shown to be important for bnMAb neutralization in the same set of 31,788 virus sequences.