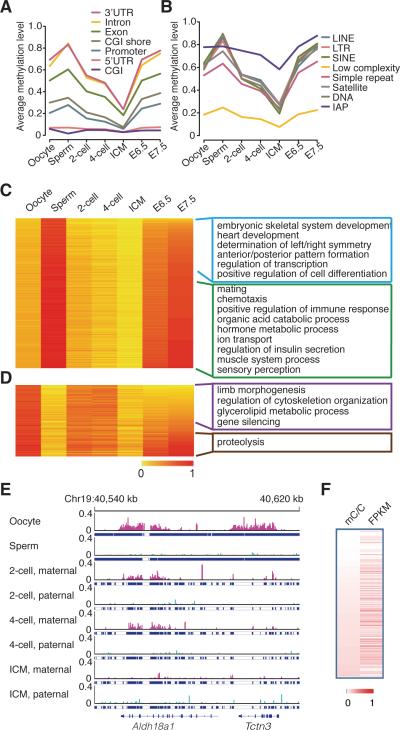
Figure 1. Dynamics of DNA methylomes for gametes and early embryos.
(A) The dynamics of methylation levels of different genomic elements (genic related classification) during early embryogenesis. The average methylation is the mean value of the methylation levels of all CpGs located in the specific element.
(B) The dynamics of methylation levels of different repeat elements. IAP is the sub-category of LTR.
(C) and (D) Heat map of the methylation reprogramming of oocyte-specific hypomethylated promoters and sperm-specific hypo-methylated promoters during early embryogenesis respectively. GO term enrichment in genes with E7.5 hypomethylated promoters or hyper methylated promoters. DNA methylation level is colored from orange to red to indicate low to high.
(E) Graphical representation of a genomic region showing the methylation level of non-CpGs for tracked maternal DNA (pink) and paternal DNA (light Blue) separately. Tracked non-CG cytosines are highlighted by short blue lines. “pa” means paternal DNA; “ma” means maternal DNA.
(F) Positive-correlations between gene expression and methylated non-CG cytosines in the genic regions. Pearson correlation coefficiency is 0.20 (p value < 10−5). Each row represents one gene from the total 22,742 expressed genes. mC/C means the density of methylated non-CG cytosines. FPKM represents gene expression level. Both DNA methylation density and gene expression are colored from white to red to indicate low to high.
See also Figure S1 and S2, Table S1, S2 and S3.