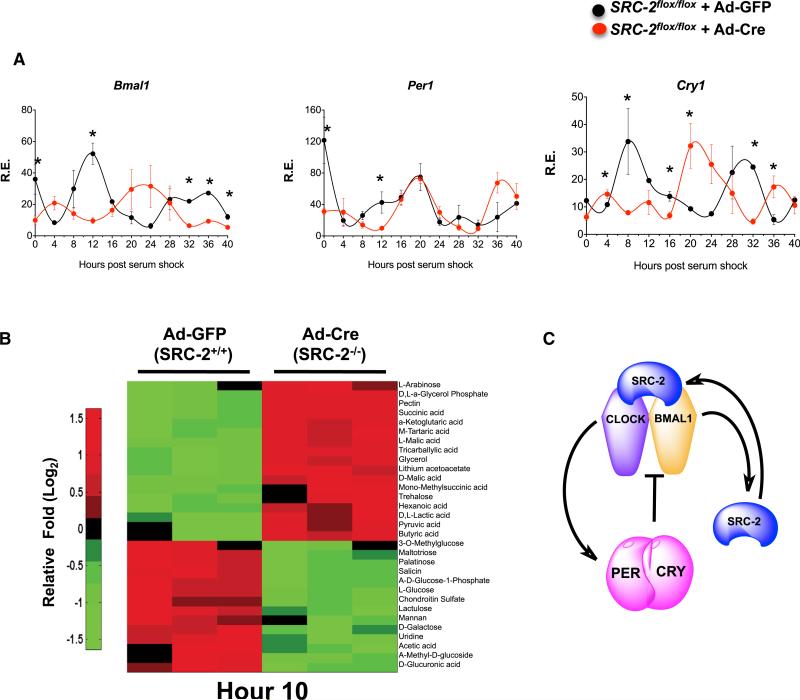
Figure 6. SRC-2 Deletion Significantly Disrupts the Cell-Autonomous Clock.
(A) qPCR analysis of circadian gene expression in synchronized MEFs isolated from SRC-2flox/flox mice infected with either Ad-GFP or Ad-Cre-GFP. Data represent triplicates at each time point for maximal gene expression of Ad-GFP normalized to one, graphed as the mean ± SEM. *p < 0.05 versus WT.
(B) Heatmap of metabolic phenotype microarray of carbon metabolism in synchronized MEFs isolated from SRC-2flox/flox mice infected with either Ad-GFP or Ad-Cre-GFP at Hr10 (n = 3). Data are graphed as the mean ± SEM. *p < 0.05 versus WT mice.
(C) Schematic representation of the role of SRC-2 as a BMAL1:CLOCK transcriptional coactivator for core clock genes.