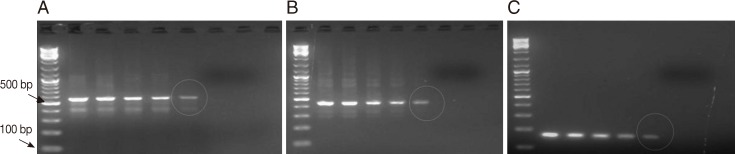
Fig. 3.
Representative ethidium bromide-stained agarose gel pictures showing PCR amplification products of protozoan DNA extracts recovered from feces seeded with various counts of oocysts/cysts by the amended kit's protocol. Encircled amplicons were the lowest number of oocysts/cysts present per extract (i.e., 200 µl) and could be detected by PCR (i.e., the lower detection limits) (A) PCR amplification products of Cryptosporidium COWP gene sequence (≈ 550 bp) using primers Cry-9/Cry-15. (B) PCR amplification products of G. lamblia gdh gene sequence (≈ 450 bp) with primers GDHeF/GDHiR. (C) PCR amplification products of E. histolytica 18S rDNA gene sequence (≈ 170 bp) with primers EntaF/EhR. Lane 1, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,700 oocysts/cysts; Lane 2, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,500 oocysts/cysts; Lane 3, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 1,000 oocysts/cysts; Lane 4, with ≈ 500 oocysts/cysts; Lane 5, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 100 oocysts/cysts; Lane 6, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 50 oocysts/cysts; Lane 7, amplification product of DNA samples retrieved from 200 µl feces spiked with ≈ 10 oocysts/cysts; M, GeneRuler™ 100 bp DNA marker.