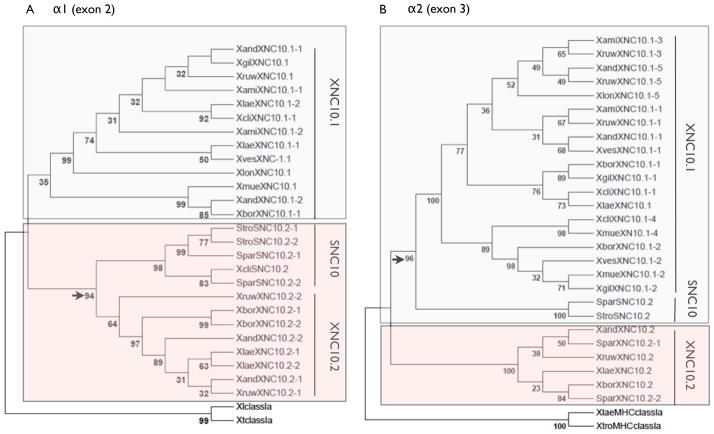
Fig 5. Phylogenetic relationships between XNC/SNC10 gene lineage within multiple species of the Xenopodinae subfamily.
Neighbor-joining trees were constructed based on amino acid alignments of (A) alpha1/exon 2 and (B) alpha 2/exon 3 of XNC/SNC10 sequences from different Xenopodinae species. Trees were rooted with X. laevis and X. tropicalis MHC class Ia genes, accession numbers: ABA43373.1 and AAP36728.1 respectively. The trees were drawn using MEGA 5.2. using pairwise gap deletions and the p-distance method to estimate evolutionary distance and confidence values were measured using 10,000 bootstrap replications with the values indicated at key nodes. Trees generated based on nucleotide alignments resulted in similar topology (data not shown).