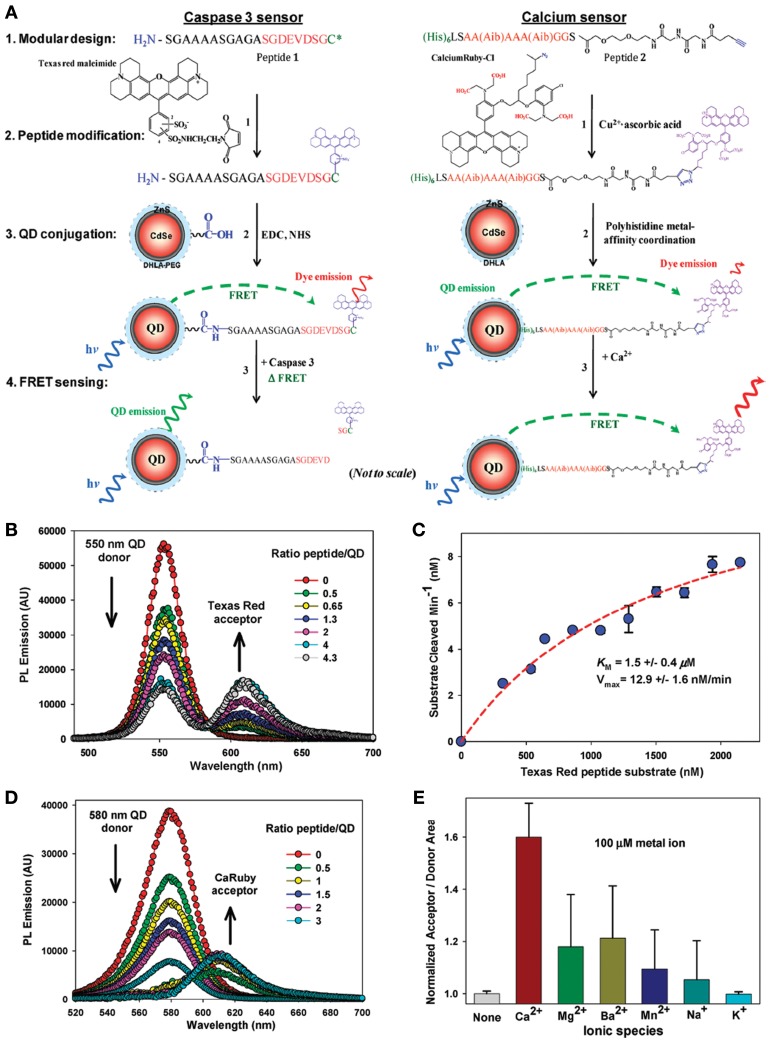
Figure 6.
Quantum dot protein biosensors. (A) Schematic showing the common design, chemical and sensing elements, including FRET-based biosensors: (1) peptide modularity, (2) peptide labeling, (3) attachment to QDs, and (4) FRET-based sensing for both the caspase 3 proteolytic sensor (left) and Ca2+ sensor (right). The 4-pendant carboxyl groups that interact with Ca2+ ions are shown in red on the CaRbCl structure. Within the Ca2+ sensor peptide sequence, Aib is the synthetic amino acid α-aminoisobutyric acid. Reactive dye structures are shown where appropriate along with the chemical linkages attaching them to the peptides. (B) Representative, superimposed spectra collected from 550 nm emitting QD donors surface functionalized with 85:15 DHLAPEG600-COOH/DHLA-PEG750-OMe ligands and covalently conjugated to increasing molar ratios of Texas Red-labeled substrate peptide. Samples excited at 350 nm. (C) Proteolytic assay data from exposing a constant concentration of 550 nm emitting QDs conjugated to 4 Texas Red substrate peptides to a constant concentration of caspase 3 enzyme. Derived Km and Vmax values are given. An R2 = 0.98 was obtained for the fitting of the curve. (D) Representative, superimposed, and deconvoluted spectra collected from 580 nm emitting QD donors self-assembled with increasing CaRbCl-acceptor labeled peptides. Samples were excited at 350 nm. (E) Normalized acceptor/donor PL area ratios for 580 nm QDs self-assembled with ~2 CaRbCl-acceptor labeled peptides exposed to selected ionic materials. The ratio from the native unexposed sensor was set to an initial value of 1 for comparison purposes (Prasuhn et al., 2010). Reproduced with permission from Prasuhn et al. (2010), Copyright 2013.