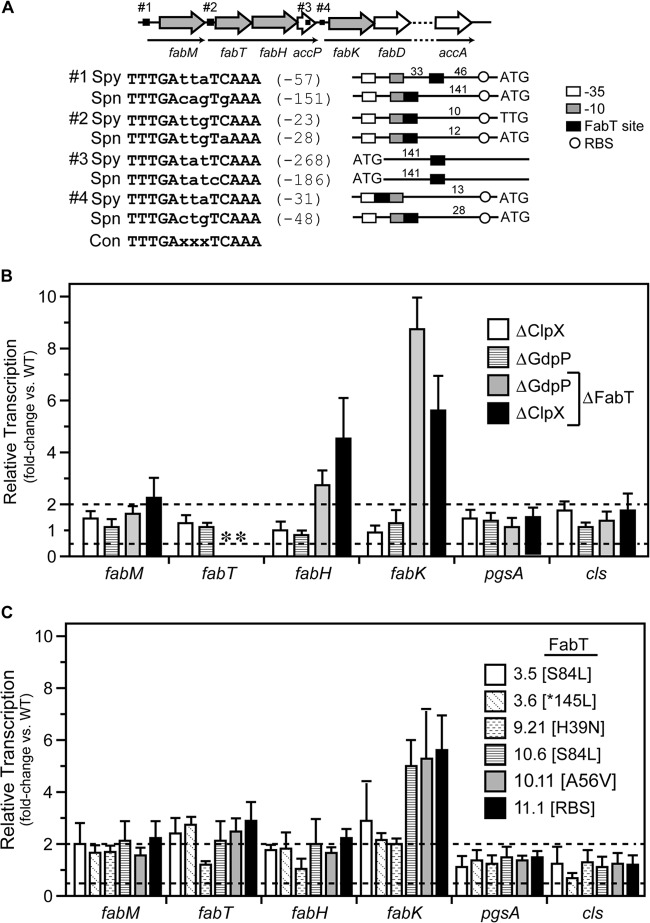
FIG 3.
Mutation of the FabT transcriptional repressor results in upregulation of fatty acid biosynthetic genes but not anionic lipid genes. (A) The fatty acid biosynthesis (fab) region carried within S. pyogenes contains 3 predicted operons (thin black arrows) and 4 putative FabT binding sites upstream (black boxes) or within fab genes (hollow arrows). Genes interrogated for mRNA expression levels are shaded in gray. Predicted FabT binding sites are shown in comparison with S. pneumoniae, and nucleotide distances in relation to translational start codons (ATG or TTG) are indicated in parentheses. Distances between FabT binding sites and predicted −35 and −10 promoter elements and the ribosome binding site (RBS) are indicated in the diagram. The consensus 13-nucleotide palindromic FabT binding site is shown (Con), and deviations are indicated in lowercase. (B and C) The relative level of message of select genes in deletion mutants (B) and PBr mutants containing FabT SNPs (C) was determined using real-time RT-PCR and normalized to the wild-type strain. Data shown are the means and the standard errors of the means derived from triplicate determinations of samples from at least three independent experiments. The dashed lines indicate >2-fold or <2-fold changes. An asterisk indicates an undetectable level due to the absence of the appropriate gene in the deletion mutant.