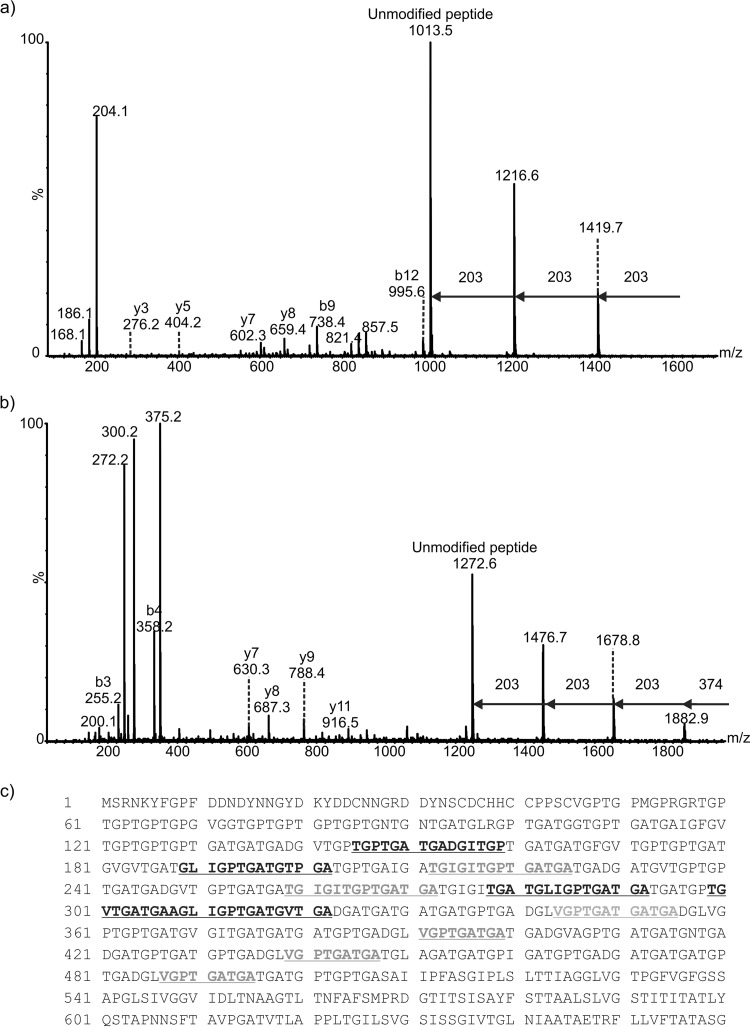
FIG 3.
Mass spectrometry analysis of peptides from proteinase K digestion of C. difficile R20291 endospore surface extracts. (a) nLC-MS/MS spectrum of the doubly protonated glycopeptide ion at m/z 811.8. Peptide type y and b ions were visible and gave the peptide sequence AGLIGPTGATGV, a peptide from the BclA3 protein. The spectrum was dominated in the high-m/z region by sequential neutral losses of 203 Da, with the unmodified peptide ion observed at m/z 1,013.5. Combined with the observed intense glycan oxonium ion at m/z 204 and neutral losses of water to give glycan-related ions at m/z 186 and 168, this spectrum suggested the peptide was modified with a chain of 3 HexNAc moieties. (b) nLC-MS/MS spectrum of a doubly protonated glycopeptides ion at m/z 1,129. Peptide type y and b ions corresponded to a sequence of TGPTGATGADGITGP, corresponding to the BclA3 protein. The high-m/z region of the spectrum was dominated by sequential neutral losses of 374 Da, 203 Da, 203 Da, and 203 Da. An intense oxonium ion was observed at m/z 375, and a very weak oxonium ion was observed at m/z 204 (not indicated). Glycan-related fragment ions were observed at m/z 300 and 272. (c) Peptide sequence coverage map of BclA3 protein homolog from spore surface protein extraction (band 1). Boldface and underlining indicate peptides modified with glycan moieties. Two of the peptides shown to be modified with glycan are shown in boldface gray text to indicate that the amino acid sequence appears in the BclA3 protein more than once. Dotted underlining indicates a glycopeptide sequence that is common to both BclA3 and BclA2 proteins.