FIG 1.
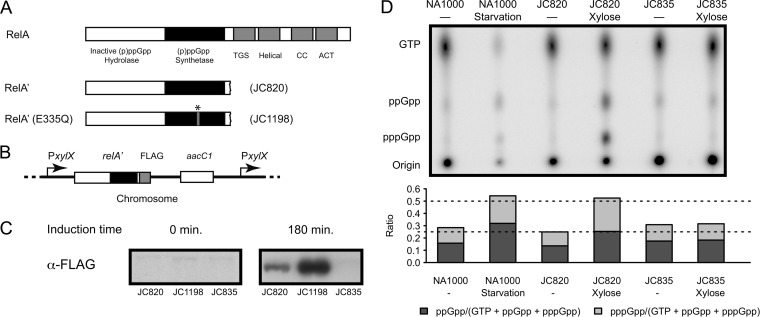
C. crescentus cells expressing RelA′-FLAG accumulate high levels of (p)ppGpp. (A) Schematics showing the organization of the different domains of the E. coli RelA protein and of the truncated RelA′ and RelA′(E335Q) proteins used in the present study. The TGS/CC/ACT domains form the regulatory region of the protein (3, 7). The star indicates the E335Q mutation. (B) Genetic constructs created to express RelA′-FLAG and RelA′(E335Q)-FLAG in C. crescentus. Plasmid pXTCYC-4-relA′-FLAG or pXTCYC-4-relA′(E335Q)-FLAG was integrated at the native xylX promoter in the C. crescentus chromosome, giving strains JC820 and JC1198, respectively. (C) RelA′-FLAG and RelA′(Q335A)-FLAG proteins are expressed in the JC820 and JC1198 strains, respectively, upon xylose addition in rich medium. JC820, JC1198 and JC835 (control strain) strains were cultivated in exponential phase in PYEG medium, and 0.3% xylose was added (PYEGX) at time zero. Cell extracts collected at time zero and 180 min after xylose addition were used to perform immunoblotting experiments using anti-FLAG antibodies. (D) Intracellular levels of (p)ppGpp in RelA′-FLAG-expressing cells, compared to starved wild-type cells. Strains NA1000 (wild-type strain), JC835 and JC820 were cultivated for 2.5 h in M5GG medium with or without 0.3% xylose. When indicated, NA1000 cells were starved in M5 medium for 90 min. The TLC autoradiograph image shown in the upper part of the figure was used to calculate the pppGpp/(GTP+ppGpp+pppGpp) and ppGpp/(GTP+ppGpp+pppGpp) ratios shown in the lower panel.