FIG 6.
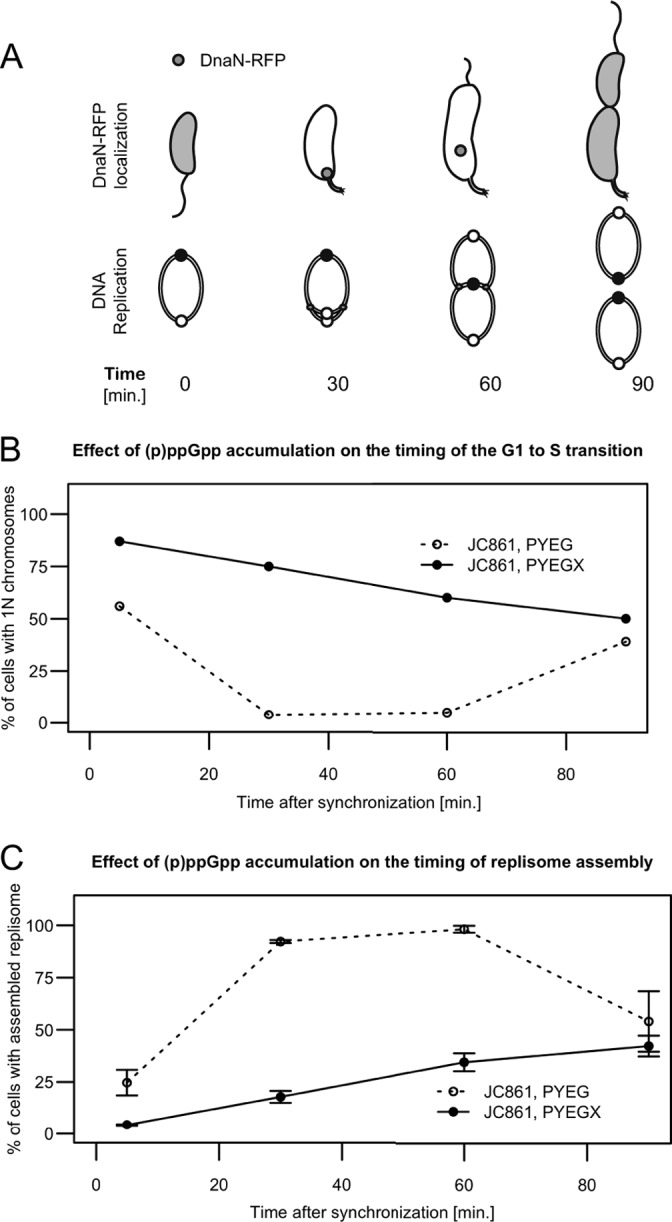
The accumulation of (p)ppGpp delays the initiation of chromosome replication. (A) Schematics showing the status of chromosome replication as a function of time when swarmer cells are grown in rich medium. The upper schematic shows that DnaN-RFP localizes as a moving focus only when DNA replication is ongoing. The lower schematic shows the replicating circular chromosome of C. crescentus as a function of the cell cycle. The small white and black circles on the lower schematic indicate the chromosomal origin and terminus, respectively. (B) (p)ppGpp accumulation delays the initiation of chromosome replication in swarmer cells. Quantification of flow cytometry experiments using isolated swarmer cells from strain JC861 (NA1000 dnaN::dnaN-RFP pXTCYC-4-relA′-FLAG) grown in PYEG or PYEGX. Cells were sampled at the indicated times, treated with rifampin, and then fixed and stained prior to flow cytometry analysis. The graph shows the percentage of cells with 1N chromosome, corresponding to cells that had not initiated chromosome replication at the indicated times of the cell cycle. (C) (p)ppGpp accumulation delays the assembly of the replisome in swarmer cells. Quantification of fluorescence microscopy experiments using isolated swarmer cells from strain JC861 grown in PYEG or PYEGX. Typical images acquired at different times under each condition and used for this quantification are shown in Fig. S8 in the supplemental material. The graph shows the percentage of cells with visible DnaN-RFP fluorescent foci, corresponding to cells that had initiated DNA replication at the indicated times of the cell cycle. The plotted values are the average percentages over three microscope fields; error bars indicate the standard deviations.
