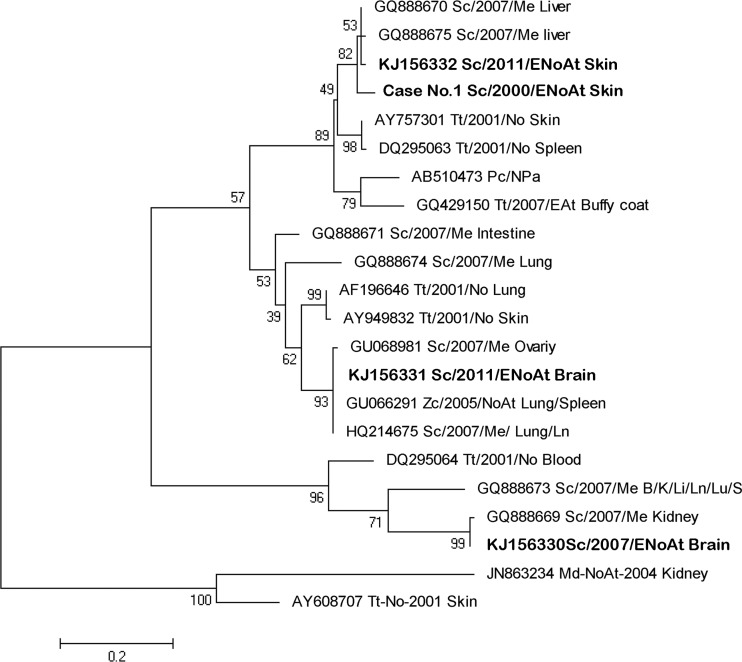
FIG 4.
Neighbor-joining phylogram of 22 selected sequences from the polymerase gene of a cetacean alphaherpesvirus. MEGA 5.0 was used to construct the maximum likelihood phylogenetic trees. A Tamura-Nei substitution model and a bootstrap resampling (1,000 replicates) were used to assess the reliability of the tree. The new isolates from this study are shown in bold type. The scale bar shows the number of nucleotide substitutions per site. The name of each sequence includes the GenBank accession number (when available); cetacean species (Md; Mesoplodon densirostris; Me; Mesoplodon europaeus; Pc; Pseudorca crassidens; Sc, S. coeruleoalba Tt, T. truncatus; Zc; Ziphius cavirostris), the year and the geographic area of the stranding (At, Atlantic Ocean; EnoAt, Northeast Atlantic Ocean; EAt, East Atlantic Ocean; Me, Mediterranean Sea; No, North Sea; NoAt, North Atlantic Ocean; Npa; North Pacific Ocean; Pa, Pacific Ocean), and the tissues in which they were detected (B, brain; I, intestine; K, kidney; Li, liver; Ln, lymph node; Lu, lung; ovary; skin; S, spleen).