Abstract
Fungal infections in the clinic have become increasingly serious. In many cases, the identification of clinically relevant fungi remains time-consuming and may also be unreliable. Matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) is a newly developed diagnostic tool that is increasingly being employed to rapidly and accurately identify clinical pathogenic microorganisms. The present meta-analysis aimed to systematically evaluate the accuracy of MALDI-TOF MS for the identification of clinical pathogenic fungi. After a rigorous selection process, 33 articles, involving 38 trials and a total of 9,977 fungal isolates, were included in the meta-analysis. The random-effects pooled identification accuracy of MALDI-TOF MS increased from 0.955 (95% confidence interval [CI], 0.939 to 0.969) at the species level to 0.977 (95% CI, 0.955 to 0.993) at the genus level (P < 0.001; χ2 = 15.452). Subgroup analyses were performed at the species level for several categories, including strain, source of strain, system, system database, and modified outcomes, to calculate the accuracy and to investigate heterogeneity. These analyses revealed significant differences between the overall meta-analysis and some of the subanalyses. In parallel, significant differences in heterogeneity among different systems and among different methods for calculating the identification ratios were found by multivariate metaregression, but none of the factors, except for the moderator of outcome, was significantly associated with heterogeneity by univariate metaregression. In summary, the MALDI-TOF MS method is highly accurate for the identification of clinically pathogenic fungi; future studies should analyze the comprehensive capability of this technology for clinical diagnostic microbiology.
INTRODUCTION
Pathogenic fungi have been increasingly detected in clinical microbiological laboratories in recent years. Invasive fungi have received growing attention for their potentially life-threatening pathogenicity (1), especially in patients undergoing transplants or receiving treatment for malignancies (2). Therefore, the development of precise, rapid, and cost-effective methods to identify clinically relevant fungi appears crucial. Unfortunately, the identification of yeast and yeast-like fungi, as well as filamentous fungi, in many clinical laboratories still mainly depends on phenotypic or molecular methods that are time-consuming, labor-intensive, and often inconclusive (3–5). As an alternative to these standard identification methods, the rapid, cost-effective, and accurate (6, 7) method matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) has been widely used in recent years in European clinical microbiology laboratories for the identification of bacteria, mycobacteria, and fungi (8, 9). The technology is now being adopted in clinical laboratories worldwide (10) due to its superiority over traditional methods for some tasks. The clearance of the Vitek MS, a product of bioMérieux, for clinical use by the State Food and Drug Administration (SFDA) of China in 2012 and by the U.S. FDA in 2013 (11) paves the way for many more clinical laboratories in these two countries to adopt MALDI-TOF MS to identify clinical pathogens. Because of its promise, some experts even describe MALDI-TOF MS technology as “a revolution in clinical microbiology” (12). Currently, four commercial systems are in use worldwide: the MALDI Biotyper (Bruker Daltonics, Bremen, Germany), the Saramis (AnagnosTec, Potsdam, Germany), and, more recently, the Andromas (Andromas, Paris, France) and Vitek MS (bioMérieux, Marcy l'Etoile, France) systems (13). Each of the four systems is equipped with a MALDI-TOF MS instrument from either Bruker Daltonics or Shimadzu (9). Among them, the MALDI Biotyper is commonly used in conjunction with an instrument from the same manufacturer. The Saramis, which is mainly combined with an Axima instrument (a product of Shimadzu), was purchased by bioMérieux in 2010 to be incorporated into the Vitek MS series (10). Finally, the Andromas system, including three distinct databases, which can be used in conjunction with either Bruker or Shimadzu instruments, was primarily used in France (8, 10). Although many articles have reported bacterial identification by MALDI-TOF MS and have compared available systems (14–16), studies comparing or using MALDI-TOF MS systems for the identification of clinically related fungi have been relatively rare. In addition, many studies have included only a few strains, and some results have been inconsistent (17, 18). Therefore, the present work aimed to analyze the gross accuracy of MALDI-TOF MS using different systems to identify clinically pathogenic fungi by performing a meta-analysis that synthesizes large amounts of data to improve the reliability of the results.
MATERIALS AND METHODS
Search strategy.
We queried PubMed (up to 18 October 2013) with the string “(maldi-ms [MeSH Terms] AND fungi [MeSH Terms]) AND (identification [Title/Abstract] OR detection [Title/Abstract])” to identify relevant articles. The Embase database was also searched (before 23 October 2013) with the words “maldi,” “fungi,” “identification,” and “detection.” The language, publication status, and geographical distribution of the publications were not restricted. In addition, we scanned the references of the eligible studies and reviews that were identified. The authors of the original studies were contacted for detailed information if the full text could not be obtained from the database. The meta-analysis was performed by referring to (when appropriate) the PRISMA guidelines (19). EndNote X4 (Thomson Reuters) was used for literature management.
Two investigators (H.L. and J.S.) independently performed the literature search and data extraction. Disagreements were resolved by discussion and/or consultation with a third researcher (Z.W.).
Study selection criteria and data extraction.
Studies evaluating the accuracy of MALDI-TOF MS for identification of clinical fungi by comparison with reference methods were considered eligible for the meta-analysis. Articles relating to the validation of the MALDI-TOF MS fungal database expanded by researchers or to the evaluation of commercial databases were included. Data on the identification accuracy of MALDI-TOF MS for fresh/frozen clinical isolates and isolates previously confirmed by gold standard methods were all included.
Studies or data were excluded if they fell into the following categories: studies with no abstract; studies on technological innovations, such as modification of the preanalytical steps of MALDI-TOF MS identification; studies applying MALDI-TOF MS to the identification of industrial/environmental microorganisms or plant- or animal-pathogenic microbes; studies using MALDI-TOF MS for the identification of clinical nonfungal pathogens or studies identifying clinical pathogens by mass spectroscopy methods other than MALDI-TOF; studies with fewer than 40 specimens, with reference methods that did not include molecular biology, or lacking a comparator method or gold standard; studies on drug resistance; and case reports or reviews.
Data abstraction was conducted using the numbers of total isolates and of isolates correctly identified at the genus and species levels in comparison to reference methods, when pertinent data were available. Data were separately collected according to the category of strain, the MS system used (Andromas, Biotyper, Saramis, Vitek MS), and the calculation method used for identification ratios (whether those data were excluded in the absence of referential mass spectral entries). In addition, the following information was abstracted: the first author, publication year, study design (prospective or retrospective), source of strains (clinical isolates only or reference strains added), database of system (commercial database only or self-established database added), threshold, geographical distribution of strains, blinded status, and reference methods. When two or more thresholds were included in a study, the one by which more fungi could be correctly identified, compared to the reference method, was abstracted.
Quality assessment.
The following modified criteria, referring to the quality assessment for studies of diagnostic accuracy (QUADAS) (20), were used to assess the quality of original studies: study design, category and geographical distribution of strains, blinded status, reference methods, threshold, strain source, and system database.
Statistical analysis.
The major effect-size index consisted of the correct identification ratios. Specifically, the identification ratios obtained after MALDI-TOF MS identification were compared to results obtained with the reference method used in each of the studies, as some researchers have found that using a lower threshold can in many cases provide a higher identification ratio, compared to the reference method, than that specified by the manufacturer (21, 22). The identification ratio was calculated as the number of correctly identified isolates divided by the total number of isolates. Before the synthesis, the data were preprocessed. To obtain the overall performance of MALDI-TOF MS for accurate identification of clinical fungal isolates, we averaged the both identification ratios when two identification systems were employed to identify the same strains in a study. We then split the identification ratios on different systems to analyze the identification performance of an individual system. Given that the data to be analyzed were nonnormally distributed, double arcsine transformation (23) was implemented before data synthesis. The transformation results in a roughly normally distributed variable (24, 25) and makes the variance more stable than the “canonical” logit transformation for ratios (24, 26).
The double arcsine-transformed ratios were subsequently pooled in fixed-effect and random-effects models (27). To better understand the results, pooled transformed estimate formulas were back-transformed into the “original units” of ratios (25). The random-effects model pooled ratios were adopted when significant heterogeneity was present. Otherwise, fixed-effect model pooled ratios were adopted. Subgroup analyses at the species level were performed on the following categories: strain, source of strain, system, and system database. In addition, the highest correct identification ratios (data were excluded in the absence of referential mass spectral entries) performed on the four systems and on the Biotyper were also analyzed separately, as most uncommon fungal species can in principle be correctly identified by the enrichment of reference spectra in the library. Some exceptions include certain species for which satisfactory spectra are inherently difficult to obtain, such as Candida guilliermondii or Cryptococcus neoformans. Comparison of identification accuracy at the genus and species levels, as well as comparisons between subanalyses with various situations and the overall species-level analysis, was performed using a Pearson chi-square test.
To analyze possible sources of high-level heterogeneity and to validate the effects of subanalyses, multivariate and univariate metaregressions were performed (27, 28) with the default DerSimonian-Laird method (29). Interpretive parameters of univariate metaregressions were provided to enable comparisons across identifications. An influence (sensitivity) analysis (29) with the random-effects model for the enrolled articles at the species level was undertaken to inspect the influence of individual research on the overall identification accuracy.
Heterogeneity between studies was estimated with Cochran's Q statistic and the I2 measure. Publication bias was evaluated by using the rank correlation method of Begg and Egger's regression method (29). Statistical significance was defined as a P value of <0.1 and an I2 value of <50% for the qualitative Cochran's Q and quantitative I2 measures, respectively. For metaregression, for the Pearson chi-square test and for publication bias interpretations, statistical significance was set as a P value of <0.05. All analyses were performed with R statistical software (version 3.0.2; R Foundation for Statistical Computing, Vienna, Austria) and with the analysis packages meta (version 3.1-2) and metafor (version 1.9-2).
RESULTS
Eligible studies.
A total of 282 items were obtained by searching PubMed with defined retrieval strings. After additional Embase retrieval and duplicate removal, 2 other citations were included. A total of 242 articles were excluded after title and/or abstract review. Among the excluded articles, 2 were excluded because no abstract was provided; 1 case report and 1 report with fewer than 40 strains were excluded; 4 were excluded due to their relating to drug resistance; 15 were excluded due to their focus on technological innovation; 130 were excluded because they reported the use of MALDI-TOF MS in non-clinically related research; 35 were rejected due to a focus on the application of MALDI-TOF to clinically related research other than the identification of clinical fungi; and 27 were discarded because they reported the use of a mass spectrometry technique other than MALDI-TOF or because they had no relationship with both MALDI-TOF and the identification of clinical fungi. In addition, 11 were excluded because they did not focus on identification accuracy despite being related to the identification of clinical fungi by MALDI-TOF, and 1 was rejected because it was not feasible to translate the article into English. After the papers were screened, 42 citations remained for full-text examination. Another 9 studies were excluded after full-text scanning, as they did not meet the inclusion criteria as described (e.g., the absence of a reference method or the absence of molecular biology). As a result, 33 articles involving 38 trials were included in this meta-analysis. Among these articles, 4 (7, 8, 13, 18) reported on the identification performance of the Andromas system, 24 (1, 4, 13, 17, 21, 22, 30–47) reported on the identification performance of the Biotyper system, 6 (21, 30, 42, 48–50) reported on the identification performance of the Saramis system, and 4 (36, 51–53) reported on the identification performance of the Vitek MS system. Notably, 5 were comparison studies between two systems (three were between the Biotyper and the Saramis, one was between the Biotyper and the Andromas, and another was between the Biotyper and the Vitek MS).
Quality of studies.
The major characteristics of the enrolled eligible studies are listed in Table 1. Among the 33 enrolled studies, 16 (48.48%) were prospective, 12 (36.36%) were retrospective, and 5 (15.15%) included both types of data. Four (12.12%) studies (22, 39, 41, 49) included reference strains, while the remaining studies (87.88%) used all clinical isolates. Eight (24.24%) studies (4, 7, 18, 31, 34, 35, 41, 44) expanded the database by establishing reference spectra for clinical fungi, while others (75.76%) used the databases provided by the instrument suppliers. The strains analyzed in most of the studies (66.67%, or 22/33) were isolated from Europe. Among them, 8 studies used strains isolated from France and 2 used isolates from multiple European countries; 5 (15.15%) used isolates from America and 4 (12.12%) used isolates from Asia; among the rest, one used isolates from 3 countries on different continents and another one used isolates from the ARTEMIS and SENTRY programs involving 22 countries. Only 8 (24.24%) articles clearly stated that a blind method was used throughout the studies; one indicated that a blind method was used for the prospective part of their investigation; the others did not indicate whether a blind method was used for their studies. Twenty-seven studies were focused on the identification of yeasts, and 8 focused on mold identification. Five out of 33 articles (31, 32, 39, 48, 51) did not report thresholds for identification. The majority of the included studies (81.82%, or 27/33) employed two or more methods that acted as references. DNA sequencing of all isolates was performed in only 3 of the 5 studies employing molecular methods as a reference (22, 35, 52), despite the fact that molecular methods are considered the gold standard for the identification of pathogens.
TABLE 1.
Characteristics of the 33 reports enrolled in this meta-analysis
| Study | Study design | Organism(s) | Geographical distribution of strains | Blinded statusa | Reference method(s)b |
|---|---|---|---|---|---|
| Marinach-Patrice et al., 2009 (37) | Retrospective | Molds | France and Belgium | NR | MO and MB |
| Marklein et al., 2009 (38) | Retrospective plus prospective | Yeasts | Germany | NR | MO, BI, and MB |
| Stevenson et al., 2010 (44) | Retrospective | Yeasts | USA | NR | BI and MB |
| van Veen et al, 2010 (45) | Retrospective plus prospective | Yeasts | Netherlands | NR | BI and MB |
| Alanio et al., 2011 (7) | Prospective | Aspergillus | France | Yes | MO and MB |
| Bader et al., 2011 (30) | Prospective | Yeasts | Germany | Yes | MO, BI, and MB |
| Cassagne et al., 2011 (31) | Prospective | Molds | France | NR | MO and MB |
| Dhiman et al., 2011 (33) | Retrospective plus prospective | Yeasts | USA | Yes | BI and MB |
| Martinez-Lamas et al., 2011 (50) | Retrospective | Candida | Spain | NR | BI and MB |
| McTaggart et al., 2011 (22) | Retrospective | Cryptococcus and non-Cryptococcus yeasts | Canada | NR | MB |
| Pinto et al., 2011 (40) | Retrospective plus prospective | Candida and non-Candida yeasts | Australia | Yes for prospective part | BI and MB |
| Putignani et al., 2011 (1) | Retrospective plus prospective | Candida and non-Candida yeasts | Italy | NR | BI and MB |
| Yan et al., 2011 (47) | Prospective | Yeasts | USA | Yes | MO, BI, and MB |
| Alshawa et al., 2012 (18) | Prospective | Molds | France | Yes | MO and MB |
| Bille et al., 2012 (8) | Prospective | Aspergillus and yeasts | France | NR | BI and MB |
| De Carolis et al., 2012 (4) | Prospective | Molds | Italy | Yes | MO and MB |
| Firacative et al., 2012 (34) | Prospective | Cryptococcus | Australia | NR | MB |
| Iriart et al., 2012 (51) | Prospective | Aspergillus and yeasts | France | NR | MO, BI, and MB |
| Posteraro et al., 2012 (41) | Retrospective | Cryptococcus | Italy | NR | MB |
| Quiles-Melero et al., 2012 (39) | Retrospective | Candida | Spain | NR | BI and MB |
| Seyfarth et al., 2012 (49) | Retrospective | Yeasts | Germany | NR | BI and MB |
| Yaman et al., 2012 (46) | Prospective | Candida | Turkey | NR | BI and MB |
| Castanheira et al., 2013 (32) | Retrospective | Candida | 22 countries | NR | BI and MB |
| Ferreira et al., 2013 (17) | Retrospective | Yeasts | Spain | NR | MO, BI, and MB |
| Kolecka et al., 2014 (35) | Prospective | Malassezia yeasts | Greece, Italy and Sweden | NR | MB |
| Lacroix et al., 2014 (13) | Prospective | Candida | France | Yes | BI and MB |
| Lohmann et al., 2013 (21) | Prospective | Yeasts | France | NR | MO, BI, and MB |
| Mancini et al., 2013 (36) | Retrospective | Candida, Candida-related and non-Candida yeasts | Italy | Yes | BI and MB |
| Nenoff et al., 2013 (48) | Retrospective | Molds | Germany, Uganda and China | NR | MO and MB |
| Rosenvinge et al., 2013 (42) | Retrospective | Yeasts | Denmark | NR | BI and MB |
| Sendid et al., 2013 (43) | Prospective | Yeasts | France | NR | MO, BI, and MB |
| Westblade et al., 2013 (52) | Prospective | Candida and non-Candida yeasts | North America | NR | MB |
| Won et al., 2013 (53) | Prospective | Yeasts | South Korea | NR | BI and MB |
NR, no report.
MO, morphology; MB, molecular biology; BI, biochemistry.
Overall meta-analysis.
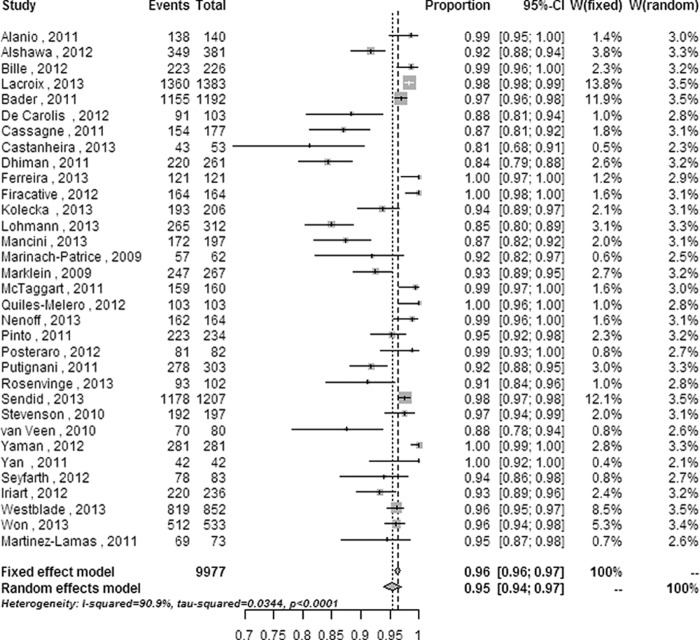
In the 33 enrolled studies, a total of 9,977 fungal isolates (8,842 yeast isolates and 1,135 mold isolates) were included. Forest plots of random-effects and fixed-effect models were used to summarize the overall statistical results of the meta-analysis at the genus and species levels (Fig. 1 and 2).
FIG 1.
Forest plot for the meta-analysis of the overall identification ratio at the genus level. CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications. Gray squares represent the weight of individual studies with the fixed-effect model; horizontal lines through the squares represent 95% confidence intervals; gray diamonds represent the overall estimate and its confidence interval; dotted vertical lines represent the fixed-effect model; and dashed vertical lines represent the random-effects model.
FIG 2.
Forest plot for the meta-analysis of the overall identification ratio at the species level. CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications.
The overall correct identification ratios of MALDI-TOF MS for clinical pathogenic fungi ranged from 0.91 to 1.00 at the genus level and from 0.81 to 1.00 at the species level. Moderate heterogeneity was found at the genus level (Q = 18.10 [P = 0.003]; I2 = 72.4% [95% CI = 36.3% to 88.0%]), and significant heterogeneity was found at the species level (Q = 352.36 [P < 0.0001]; I2 = 90.9% [95% CI = 88.3% to 92.9%]). The random-effects pooled identification accuracy of MALDI-TOF MS increased from 0.955 with a 95% CI of 0.939 to 0.969 at the species level to 0.977 with a 95% CI of 0.955 to 0.993 at the genus level (P < 0.001 and χ2 = 15.452).
Subgroup meta-analyses and investigation of heterogeneity.
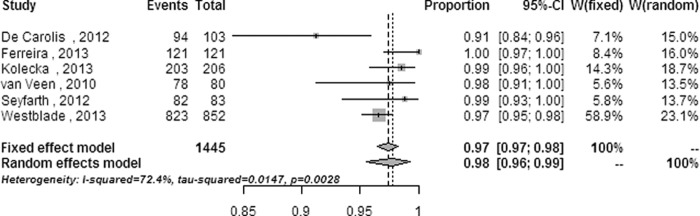
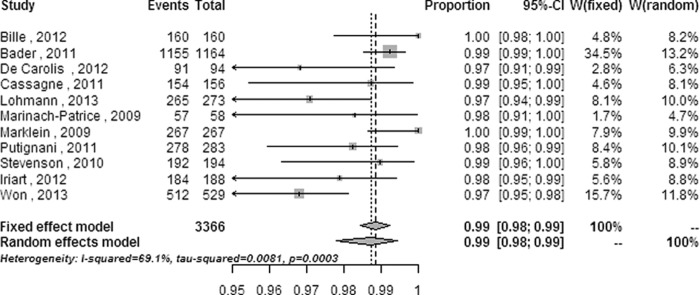
The heterogeneity and random-effects or fixed-effect pooled ratios of subgroup analyses performed at the species level on categories including strain (see Fig. S1 in the supplemental material), strain source (clinical isolates only or reference strains also) (see Fig. S2 in the supplemental material), system database (commercial database only or self-established database also) (see Fig. S3 in the supplemental material), system (see Fig. S4 in the supplemental material,) and modified outcomes (namely, the highest correct identification ratios defined in the statistical analysis) from all systems and the Biotyper (Fig. 3 and 4) are listed in Table 2.
FIG 3.
Forest plot for the subanalysis of modified outcomes from all systems at the species level. CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications.
FIG 4.
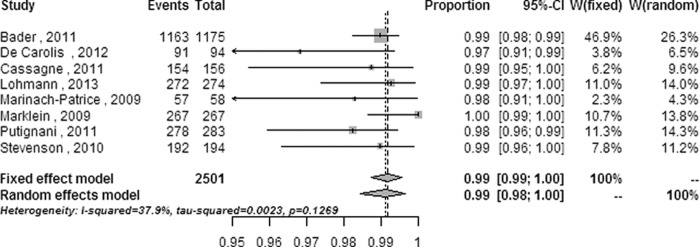
Forest plot for the subanalysis of modified outcomes from the Biotyper system at the species level. CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications.
TABLE 2.
The heterogeneity and pooled correct identification ratios by subgroup analyses
| Subanalysis | No. of isolates (no. of studies) | Within-group heterogeneity |
Correct identification ratio (95% CI)a | P comparison with overall ratio (χ2 value) | |
|---|---|---|---|---|---|
| P (Q value) | I2 (95% CI [%]) | ||||
| Category of strains | |||||
| Yeasts | 8,842 (27) | <0.0001 (291.38) | 91.1 (88.2–93.2) | 0.959 (0.943–0.973) | 0.129 (2.309) |
| Molds | 1,135 (8) | <0.0001 (45.71) | 84.7 (71.6–91.7) | 0.934 (0.888–0.969) | 0.002 (9.961) |
| Systems | |||||
| Andromas | 2,130 (4) | <0.0001 (33.68) | 91.1 (80.3–96.0) | 0.972 (0.936–0.994) | <0.001 (13.202) |
| Biotyper | 7,289 (24) | <0.0001 (303.10) | 92.4 (89.9–94.3) | 0.954 (0.933–0.971) | 0.725 (0.124) |
| Saramis | 1,926 (6) | <0.0001 (63.71) | 92.2 (85.7–95.7) | 0.938 (0.881–0.978) | 0.001 (10.477) |
| Vitek MS | 1,818 (4) | <0.0001 (31.79) | 90.6 (78.9–95.8) | 0.933 (0.887–0.968) | <0.001 (16.923) |
| Source of strains | |||||
| Clinical isolates only | 9,549 (29) | <0.0001 (332.42) | 91.6 (89.0–93.5) | 0.950 (0.932–0.965) | 0.097 (2.755) |
| Clinical isolates plus reference strains | 428 (4) | 0.0304 (8.92) | 66.4 (1.5–88.5) | 0.988 (0.960–1.000) | 0.001 (10.962) |
| System database | |||||
| Commercial database only | 8,527 (25) | <0.0001 (282.37) | 91.5 (88.7–93.6) | 0.955 (0.937–0.970) | 0.988 (0) |
| Commercial database plus self-established database | 1,450 (8) | <0.0001 (63.44) | 89.0 (80.6–93.7) | 0.955 (0.916–0.983) | 0.962 (0.002) |
| Modified outcomes | |||||
| All systems | 3,366 (11) | 0.0003 (32.41) | 69.1 (42.4–83.5) | 0.987 (0.978–0.994) | <0.001 (73.927) |
| Biotyper | 2,501 (8) | 0.1269 (11.28) | 37.9 (0–72.6) | 0.992b (0.987–0.995) | <0.001 (74.183) |
Random-effects pooled ratios, except where noted otherwise.
Fixed-effect pooled ratio.
The heterogeneity was not obviously decreased in subgroup meta-analyses, except in subanalyses on isolates with reference strains added and on modified outcomes performed on all systems and on the Biotyper. The heterogeneity decreased from a significant level (Q = 352.36 [P < 0.0001] and I2 = 90.9% [95% CI = 88.3% to 92.9%] for the overall meta-analysis) to a moderate level (Q = 8.92 [P = 0.0304] and I2 = 66.4% [95% CI = 1.5% to 88.5%] for subanalysis of isolates with reference strains added; Q = 32.41 [P = 0.0003] and I2 = 69.1% [95% CI = 42.4% to 83.5%] for subanalysis of modified outcomes performed on all systems) or a low level (Q = 11.28 [P = 0.1269] and I2 = 37.9% [95% CI = 0 to 72.6%] for subanalysis of modified outcomes performed on the Biotyper system).
There were significant differences in the correct identification ratios at the species level (P < 0.05 for each comparison) between the overall meta-analysis and some of the subanalyses, including mold isolates, the identification system (Andromas, Saramis, and Vitek MS systems), isolates identified in studies with reference strains added, and modified outcomes from all systems and of the Biotyper system (Table 2). No significant differences (P > 0.05 for each comparison) were observed between the overall meta-analysis and the other subanalyses. The correct identification performances of the subanalyses on the Andromas system, on isolates with reference strains added, and on modified outcomes were superior to the overall ratio, while those of the subanalyses on mold identification and on the Saramis and Vitek MS systems were inferior to the overall ratio in this meta-analysis.
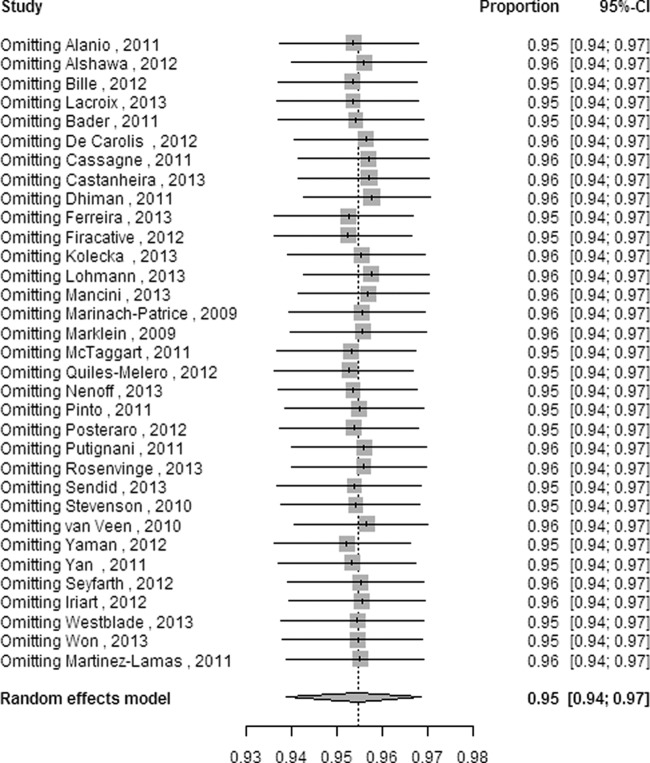
Slight significant differences (P = 0.049) in heterogeneity were found among different systems through multivariate metaregression with 4 moderators: strain (yeasts versus molds), system, source (source of strain), and database (database of system). Still, with 5 moderators (strain, system, source, database, and outcome [namely, the calculation method for identification ratios represented in the study selection criteria and data extraction]), significant differences in heterogeneity among different systems (P = 0.026) and among different calculation methods (P = 0.004) were discovered. The results of multivariate metaregression confirmed the univariate associations in all cases except for the moderator of system, which showed a significant difference by multivariate metaregression but no significant difference (P = 0.07) by univariate metaregression. None of the factors, except for the moderator of outcome, was significantly associated with heterogeneity by univariate metaregression (Table 3). Influence analysis (Fig. 5) showed that no individual study had any obvious influence on the combined overall ratio at the species level; this analysis was performed by inspecting pooled estimates that were calculated by omitting one study at a time (29).
TABLE 3.
Univariate metaregression for ratios of correct identification
| Moderator | Metaregression coefficient | 95% CI | P |
|---|---|---|---|
| Source | 0.1231 | −0.0693 to 0.3156 | 0.2099 |
| Strain | −0.0737 | −0.2091 to 0.0617 | 0.2863 |
| Database | 0.0214 | −0.1115 to 0.1543 | 0.7522 |
| System | −0.0589 | −0.1225 to 0.0048 | 0.0700 |
| Outcome | 0.1493 | 0.0382 to 0.2604 | 0.0084 |
FIG 5.
Influence analysis with a random-effects model for the enrolled articles at the species level. CI, confidence interval.
Assessment of publication bias.
Begg rank correlation (with continuity correction) and Egger's linear regression test of funnel plot asymmetry at the species level showed that little publication bias was detected in this meta-analysis (z = −0.341 and P = 0.733 for Begg; t = −1.576 and P = 0.125 for Egger's).
DISCUSSION
As a newly developed technology for the identification and antimicrobial resistance analysis of clinical pathogens (54, 55), MALDI-TOF MS has shown many merits, as described above. This meta-analysis highlights the performance of the four available MALDI-TOF MS systems for the accurate identification of clinical pathogenic fungi at both the genus and species levels. The evaluation results demonstrate that MALDI-TOF MS is a highly accurate technology for clinical fungus identification, with a correct identification proportion of 0.955 (95% CI = 0.939 to 0.969) at the species level and 0.977 (95% CI = 0.955 to 0.993) at the genus level. Among individual systems, the Andromas system yielded the highest performance in the four synthesized studies enrolled in this meta-analysis, showing an accuracy of 0.972 (95% CI = 0.936 to 0.994) at the species level. However, it is still uncertain whether the accuracy of the Andromas system for the identification of clinical fungi is generally superior to those of the other three systems. This uncertainty remains because there are currently no sufficiently direct comparisons between the Andromas system and other systems (13), and the identification ratios can be affected by many factors (for example, the category of strain, the proportion of common and unusual species, or the reference library version) that were revealed by the severe heterogeneity in the pooled ratios. It comes as no surprise that the identification accuracy of the Vitek MS was similar to that of the Saramis (0.933 [95% CI, 0.887 to 0.968] for the Vitek MS versus 0.938 [95% CI, 0.881 to 0.978] for the Saramis), as the former was developed on the basis of the latter (36).
From a professional standpoint, molds are more difficult to identify than yeasts in clinical practice (37). Still, an identification accuracy of 0.934 with a 95% CI of 0.888 to 0.969 revealed by MALDI-TOF MS in 8 pooled studies with 1,135 various mold isolates indicates that MALDI-TOF MS is a good method for the accurate and rapid identification of pathogenic molds, despite the fact that it was still a relatively low correct identification ratio compared to that for yeasts. At the same time, we noticed that the identification capability of MALDI-TOF MS against common and unusual fungal isolate species was variable. This observation is due not only to the inherent difficulty of obtaining satisfactory spectra from some species, such as Candida guilliermondii or Cryptococcus neoformans, but also to the insufficient numbers of spectra for uncommon fungal species in commercial reference libraries (at least in their previous versions). Thus, it will be increasingly important to update these libraries to continuously enrich them with fungal strains and species that are absent or poorly represented in their current versions, although there was no significant difference (P = 0.962) in the correct identification ratios at the species level between the overall meta-analysis and the subanalysis with researcher-expanded databases. Fortunately, commercial databases are continuously improved and updated at approximately 3- to 6-month intervals (12).
Subanalysis of studies in which the clinical strains were supplemented with reference strains decreased the heterogeneity to 66.4% with a 95% CI of 1.5% to 88.5% and improved the accuracy to 0.988 with a 95% CI of 0.960 to 1.000 in the four enrolled studies (428 isolates), implying that strains with substantive and distinct properties can be more easily identified by MALDI-TOF MS than those with inherently ambiguous properties. For example, only 66% of Candida guilliermondii and 50% of Cryptococcus neoformans isolates can be correctly identified by MALDI-TOF MS (33, 44). The pooled correct identification ratios were raised to 0.987 with a 95% CI of 0.978 to 0.994 and to 0.992 with a 95% CI of 0.987 to 0.995 by the subanalyses with modified outcomes from the four systems and the Biotyper system, respectively, while the heterogeneity was reduced to 69.1% with a 95% CI of 42.4% to 83.5% and 37.9% with a 95% CI of 0 to 72.6%, respectively. These results indicate that the gross identification capability of MALDI-TOF MS for clinical pathogenic fungi can be further improved by updating the databases with more mass spectra for unusual species.
Clinically, it is often not feasible to apply more elaborate, bivariate meta-analysis models to the evaluation of identification instruments for clinical pathogens due to a lack of direct comparisons; thus, the synthesis of single ratios is unavoidably adopted (27, 56). Still, the stability in a meta-analysis of single ratios is different from that of bivariate meta-analysis, and in many cases, the heterogeneity among individual ratios is very severe, as has been demonstrated in many previously published articles on single ratios (27, 28, 57). Such seemingly inherent and significant heterogeneity also appeared in this meta-analysis; nevertheless, subanalyses from several clinical aspects were performed.
Many fungal diseases have a worldwide distribution, whereas others are endemic to specific geographical regions (2). The isolates in the present enrolled studies mainly came from Europe, where MALDI-TOF MS has been used for several years, and only a few strains in one study came from Africa (48); such situations will inevitably call the identification capability of MALDI-TOF MS into question. Similarly, nearly half of the enrolled studies were retrospective, and 72.73% of studies did not clearly report a blinding method, both of which lessen the quality of the original articles. As for reference methods, although sequencing is currently the gold standard, less than 10% of studies sequenced all isolates, and most of the studies compared MALDI-TOF MS with commonly used biochemical methods and/or morphology unless a discrepancy occurred. Thus, many studies likely overestimated the accuracy of MALDI-TOF MS when the reference phenotypic system and the system under evaluation both made mistakes.
Finally, some other meaningful limitations of the present work should be acknowledged. First, with regard to proportion, the predominant fungal isolates investigated in most of the enrolled studies were commonly detected species, which are much easier to identify than unusual species (32), and this leads to overestimations of accuracy. Second, because the number of investigations directly comparing two different systems was less than 4 (for example, there was only 1 comparing the Biotyper and Vitek MS systems), sizeable heterogeneity emerged, making comparative evaluation of the identification performance between different systems unfeasible. Third, the databases are updated frequently, and newly developed databases generally have more reference spectra than older ones; this may be another cause of the severe heterogeneity. The use of more databases and terms for literature retrieval might retrieve more articles; nevertheless, little publication bias was found in the present meta-analysis.
As an entirely new technology for the clinical diagnosis of pathogenic diseases, MALDI-TOF MS has many advantages over other current methods for pathogen identification. For example, the turnaround times for the identification of clinical fungi from colony or culture to the identification result have been routinely shortened to less than 10 min with the Andromas software (8) and approximately 13 min with the Biotyper software (46), and the detection (reagent) cost has declined to $0.50 per isolate using the Biotyper system (33). Moreover, other applications of MALDI-TOF MS have been developed, such as strain typing (58), assessing drug susceptibility (59), and detecting virulence factors (60). On the other hand, MALDI-TOF MS currently has some limitations. The high cost of purchasing and maintaining the instrument is the major limitation, and it is difficult for the method to detect antimicrobial resistance or to discriminate closely related species (10).
In summary, MALDI-TOF MS showed high accuracy for the identification of clinical pathogenic fungi in the present meta-analysis. Therefore, future studies to analyze the comprehensive capability of this technology for clinical microbiology diagnostics are warranted.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ying Xu, a postgraduate studying in the United States, for reviewing the manuscript and correcting grammar.
This work was funded by the First Affiliated Hospital of Anhui Medical University (foundation number 2011KJ07), which had no role in any of the investigation or writing.
We report no conflicts of interest.
Footnotes
Published ahead of print 14 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00700-14.
REFERENCES
- 1.Putignani L, Del Chierico F, Onori M, Mancinelli L, Argentieri M, Bernaschi P, Coltella L, Lucignano B, Pansani L, Ranno S, Russo C, Urbani A, Federici G, Menichella D. 2011. MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol. Biosyst. 7:620–629. 10.1039/c0mb00138d [DOI] [PubMed] [Google Scholar]
- 2.Richardson MD, Warnock DW. 2012. Fungal infection: diagnosis and management, 4th ed. Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 3.Chen JHK, Yam W-C, Ngan AHY, Fung AMY, Woo W-L, Yan M-K, Choi GKY, Ho P-L, Cheng VCC, Yuen K-Y . 2013. Advantages of using matrix-assisted laser desorption ionization–time of flight mass spectrometry as a rapid diagnostic tool for identification of yeasts and mycobacteria in the clinical microbiological laboratory. J. Clin. Microbiol. 51:3981–3987. 10.1128/JCM.01437-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Carolis E, Posteraro B, Lass-Florl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. 2012. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18:475–484. 10.1111/j.1469-0691.2011.03599.x [DOI] [PubMed] [Google Scholar]
- 5.Del Chierico F, Masotti A, Onori M, Fiscarelli E, Mancinelli L, Ricciotti G, Alghisi F, Dimiziani L, Manetti C, Urbani A, Muraca M, Putignani L. 2012. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J. Proteomics 75:3314–3330. 10.1016/j.jprot.2012.03.048 [DOI] [PubMed] [Google Scholar]
- 6.Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol. 50:3301–3308. 10.1128/JCM.01405-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alanio A, Beretti JL, Dauphin B, Mellado E, Quesne G, Lacroix C, Amara A, Berche P, Nassif X, Bougnoux ME. 2011. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin. Microbiol. Infect. 17:750–755. 10.1111/j.1469-0691.2010.03323.x [DOI] [PubMed] [Google Scholar]
- 8.Bille E, Dauphin B, Leto J, Bougnoux ME, Beretti JL, Lotz A, Suarez S, Meyer J, Join-Lambert O, Descamps P, Grall N, Mory F, Dubreuil L, Berche P, Nassif X, Ferroni A. 2012. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. 18:1117–1125. 10.1111/j.1469-0691.2011.03688.x [DOI] [PubMed] [Google Scholar]
- 9.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Proteomics 10:151–164. 10.1586/epr.13.8 [DOI] [PubMed] [Google Scholar]
- 10.Patel R. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 57:564–572. 10.1093/cid/cit247 [DOI] [PubMed] [Google Scholar]
- 11.bioMérieux, Inc. 2013. bioMérieux announces U.S. FDA clearance for VITEK® MS, a revolutionary technology which reduces microbial identification from days to minutes reinforcing medical value of diagnostics. bioMérieux, Inc., Durham, NC [Google Scholar]
- 12.Lavigne JP, Espinal P, Dunyach-Remy CA, Messad N, Pantel A, Sotto A. 2013. Mass spectrometry: a revolution in clinical microbiology? Clin. Chem. Lab. Med. 51:257–270. 10.1515/cclm-2012-0291 [DOI] [PubMed] [Google Scholar]
- 13.Lacroix C, Gicquel A, Sendid B, Meyer J, Accoceberry I, Francois N, Morio F, Desoubeaux G, Chandenier J, Kauffmann-Lacroix C, Hennequin C, Guitard J, Nassif X, Bougnoux ME. 2014. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for the identification of Candida species. Clin. Microbiol. Infect. 20:153–158. 10.1111/1469-0691.12210 [DOI] [PubMed] [Google Scholar]
- 14.Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia L, Westblade LF, Ferraro MJ, Branda JA. 2013. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J. Clin. Microbiol. 51:2225–2231. 10.1128/JCM.00682-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alby K, Gilligan PH, Miller MB. 21 August 2013. Comparison of MALDI-TOF mass spectrometry platforms for the identification of Gram-negative rods from cystic fibrosis patients. J. Clin. Microbiol. 10.1128/JCM.01618-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175. 10.1128/JCM.01881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira L, Sánchez-Juanes F, Vega S, González MI, García M, Rodríguez S, González-Buitrago JM, Muñoz-Bellido JL. 2013. Identification of fungal clinical isolates by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Rev. Esp. Quimioter. 26:193–197 [PubMed] [Google Scholar]
- 18.Alshawa K, Beretti JL, Lacroix C, Feuilhade M, Dauphin B, Quesne G, Hassouni N, Nassif X, Bougnoux ME. 2012. Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2277–2281. 10.1128/JCM.06634-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. 2009. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. BMJ 339:b2700. 10.1136/bmj.b2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiting P, Rutjes AW, Reitsma J, Bossuyt PM, Kleijnen J. 2003. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med. Res. Methodol. 3:25. 10.1186/1471-2288-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmann C, Sabou M, Moussaoui W, Prevost G, Delarbre JM, Candolfi E, Gravet A, Letscher-Bru V. 2013. Comparison between the Biflex III-Biotyper and the Axima-SARAMIS systems for yeast identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51:1231–1236. 10.1128/JCM.03268-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. 2011. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3050–3053. 10.1128/JCM.00651-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman MF, Tukey JW. 1950. Transformations related to the angular and the square root. Ann. Math. Statist. 21:607–611. 10.1214/aoms/1177729756 [DOI] [Google Scholar]
- 24.Barendregt JJ, Doi SA, Lee YY, Norman RE, Vos T. 2013. Meta-analysis of prevalence. J. Epidemiol. Community Health 67:974-978. 10.1136/jech-2013-203104 [DOI] [PubMed] [Google Scholar]
- 25.Miller JJ. 1978. The inverse of the Freeman-Tukey double arcsine transformation. Am. Stat. 32:138 [Google Scholar]
- 26.Trikalinos TA, Trow P, Schmid CH. 2013. Simulation-based comparison of methods for meta-analysis of proportions and rates (methods research report). U.S. Agency for Healthcare Research and Quality, Department of Health and Human Services, Washington, DC: [PubMed] [Google Scholar]
- 27.Chatzigeorgiou KS, Sergentanis TN, Tsiodras S, Hamodrakas SJ, Bagos PG. 2011. Phoenix 100 versus Vitek 2 in the identification of gram-positive and gram-negative bacteria: a comprehensive meta-analysis. J. Clin. Microbiol. 49:3284–3291. 10.1128/JCM.00182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beijer U, Wolf A, Fazel S. 2012. Prevalence of tuberculosis, hepatitis C virus, and HIV in homeless people: a systematic review and meta-analysis. Lancet Infect. Dis. 12:859–870. 10.1016/S1473-3099(12)70177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarzer G. 2013. meta: meta-analysis with R. R package version 3.1-2. http://CRAN.R-project.org/package=meta
- 30.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x [DOI] [PubMed] [Google Scholar]
- 31.Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6:e28425. 10.1371/journal.pone.0028425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castanheira M, Woosley LN, Diekema DJ, Jones RN, Pfaller MA. 2013. Candida guilliermondii and other species of candida misidentified as Candida famata: assessment by Vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 51:117–124. 10.1128/JCM.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616. 10.1128/JCM.02381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firacative C, Trilles L, Meyer W. 2012. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7:e37566. 10.1371/journal.pone.0037566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolecka A, Khayhan K, Arabatzis M, Velegraki A, Kostrzewa M, Andersson A, Scheynius A, Cafarchia C, Iatta R, Montagna MT, Youngchim S, Cabañes FJ, Hoopman P, Kraak B, Groenewald M, Boekhout T. 2014. Efficient identification of Malassezia yeasts by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS). Br. J. Dermatol. 170:332–341. 10.1111/bjd.12680 [DOI] [PubMed] [Google Scholar]
- 36.Mancini N, De Carolis E, Infurnari L, Vella A, Clementi N, Vaccaro L, Ruggeri A, Posteraro B, Burioni R, Clementi M, Sanguinetti M. 2013. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry systems for identification of yeasts of medical importance. J. Clin. Microbiol. 51:2453–2457. 10.1128/JCM.00841-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marinach-Patrice C, Lethuillier A, Marly A, Brossas JY, Gene J, Symoens F, Datry A, Guarro J, Mazier D, Hennequin C. 2009. Use of mass spectrometry to identify clinical Fusarium isolates. Clin. Microbiol. Infect. 15:634–642. 10.1111/j.1469-0691.2009.02758.x [DOI] [PubMed] [Google Scholar]
- 38.Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917. 10.1128/JCM.00389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quiles-Melero I, Garcia-Rodriguez J, Gomez-Lopez A, Mingorance J. 2012. Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:67–71. 10.1007/s10096-011-1277-z [DOI] [PubMed] [Google Scholar]
- 40.Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712. 10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50:2472–2476. 10.1128/JCM.00737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenvinge FS, Dzajic E, Knudsen E, Malig S, Andersen LB, Lovig A, Arendrup MC, Jensen TG, Gahrn-Hansen B, Kemp M. 2013. Performance of matrix-assisted laser desorption-time of flight mass spectrometry for identification of clinical yeast isolates. Mycoses 56:229–235. 10.1111/myc.12000 [DOI] [PubMed] [Google Scholar]
- 43.Sendid B, Ducoroy P, Francois N, Lucchi G, Spinali S, Vagner O, Damiens S, Bonnin A, Poulain D, Dalle F. 2013. Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med. Mycol. 51:25–32. 10.3109/13693786.2012.693631 [DOI] [PubMed] [Google Scholar]
- 44.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486. 10.1128/JCM.00687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907. 10.1128/JCM.02071-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaman G, Akyar I, Can S. 2012. Evaluation of the MALDI TOF-MS method for identification of Candida strains isolated from blood cultures. Diagn. Microbiol. Infect. Dis. 73:65–67. 10.1016/j.diagmicrobio.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 47.Yan Y, He Y, Maier T, Quinn C, Shi G, Li H, Stratton CW, Kostrzewa M, Tang YW. 2011. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization Biotyper system. J. Clin. Microbiol. 49:2528–2532. 10.1128/JCM.00339-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Graser Y. 2013. MALDI-TOF mass spectrometry—a rapid method for the identification of dermatophyte species. Med. Mycol. 51:17–24. 10.3109/13693786.2012.685186 [DOI] [PubMed] [Google Scholar]
- 49.Seyfarth F, Wiegand C, Erhard M, Graser Y, Elsner P, Hipler UC. 2012. Identification of yeast isolated from dermatological patients by MALDI-TOF mass spectrometry. Mycoses 55:276–280. 10.1111/j.1439-0507.2011.02086.x [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Lamas L, Perez del Molino ML, Pardo F, Varela E, Regueiro BJ. 2011. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry vs conventional methods in the identification of Candida non-albicans. Enferm. Infecc. Microbiol. Clin. 29:568–572. (In Spanish.). 10.1016/j.eimc.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 51.Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. 2012. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization-time of flight system with a new time-effective strategy. J. Clin. Microbiol. 50:2107–2110. 10.1128/JCM.06713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Burnham CA. 2013. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J. Clin. Microbiol. 51:2267–2272. 10.1128/JCM.00680-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Won EJ, Shin JH, Lee K, Kim MN, Lee HS, Park YJ, Joo MY, Kim SH, Shin MG, Suh SP, Ryang DW. 2013. Accuracy of species-level identification of yeast isolates from blood cultures from 10 university hospitals in South Korea by use of the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based Vitek MS system. J. Clin. Microbiol. 51:3063–3065. 10.1128/JCM.00945-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majcherczyk PA, McKenna T, Moreillon P, Vaudaux P. 2006. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 255:233–239. 10.1111/j.1574-6968.2005.00060.x [DOI] [PubMed] [Google Scholar]
- 55.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227. 10.1128/JCM.00984-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riley RD, Thompson JR, Abrams KR. 2008. An alternative model for bivariate random-effects meta-analysis when the within-study correlations are unknown. Biostatistics 9:172–186. 10.1093/biostatistics/kxm023 [DOI] [PubMed] [Google Scholar]
- 57.Vallely A, Page A, Dias S, Siba P, Lupiwa T, Law G, Millan J, Wilson DP, Murray JM, Toole M, Kaldor JM. 2010. The prevalence of sexually transmitted infections in Papua New Guinea: a systematic review and meta-analysis PLoS One 5:e15586. 10.1371/journal.pone.0015586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sauer S, Freiwald A, Maier T, Kube M, Reinhardt R, Kostrzewa M, Geider K. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843. 10.1371/journal.pone.0002843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, Posteraro B. 2012. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol. 50:2479–2483. 10.1128/JCM.00224-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bittar F, Ouchenane Z, Smati F, Raoult D, Rolain JM. 2009. MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int. J. Antimicrob. Agents 34:467–470. 10.1016/j.ijantimicag.2009.03.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.