FIG 3.
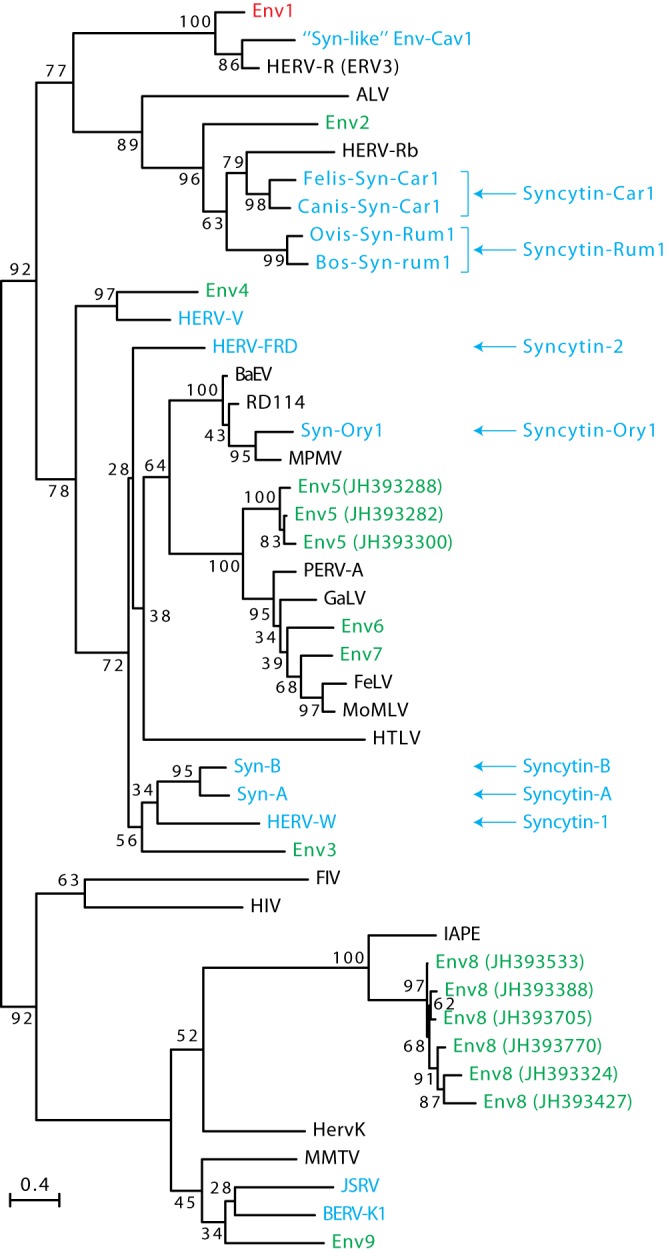
Retroviral envelope protein-based phylogenetic tree with the identified Ictidomys tridecemlineatus Env protein candidates. The maximum-likelihood tree inferred with the RaxML software was constructed using envelope amino acid sequences from mammalian endogenous retroviruses and from a series of infectious retroviruses. The horizontal branch length is proportional to the percentage of amino acid substitutions from the node (scale bar on the left), and the percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. ALV, avian leukosis virus; BaEV, baboon endogenous virus; BERV, bovine endogenous retrovirus; FeLV, feline leukemia virus; FIV, feline immunodeficiency virus; GaLV, gibbon ape leukemia virus; HERV, human endogenous retrovirus; HIV, human immunodeficiency virus; HTLV, human T cell leukemia virus; JSRV, Jaagsiekte sheep retrovirus; MMTV, murine mammary tumor virus; IAPE, intracisternal A-type particle with an envelope gene; MoMLV, Moloney murine leukemia virus; MPMV, Mason-Pfizer monkey virus; PERV, porcine endogenous retrovirus; RD114, feline endogenous type-C retrovirus.
