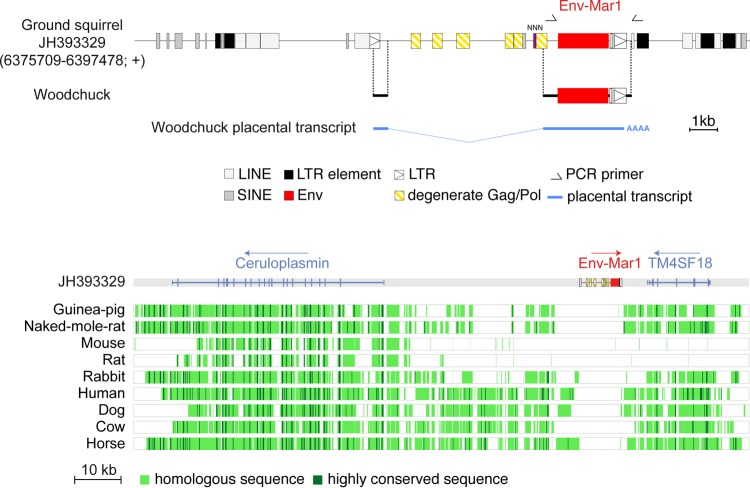
FIG 5.
Characterization of the thirteen-lined ground squirrel (Ictidomys tridecemlineatus) and woodchuck (Marmota monax) env-Mar1-containing endogenous retrovirus and of its integration site. (Upper panel) Structure of the env-Mar1-containing ERV and orthology between the ground squirrel and woodchuck env-Mar1 sequences. Repeated mobile elements (gray), proviral LTRs, and the degenerate gag-pol genes, as identified by the RepeatMasker and BLAST web programs, are positioned (the symbols used are given below the panel). Undetermined sequence is indicated by NNN. PCR primers used to identify the env-Mar1 orthologous copy in the woodchuck are indicated (black half arrows), and the regions of the determined sequences homologous to those of the ground squirrel are aligned. The spliced env subgenomic transcript as determined by 5′- and 3′-RACE of woodchuck placental RNA is indicated. (Lower panel) Absence of the env1-containing ERV in the genomes of distant mammalian lineages. The genomic locus of the Ictidomys tridecemlineatus env-Mar1-containing provirus (env in red), along with its surrounding Ceruloplasmin and TM4SF18 genes, was recovered from the UCSC Genome Browser (http://genome.ucsc.edu/), together with the syntenic loci of the indicated species genomes; the positions of exons (vertical lines) of the resident Ceruloplasmin and TM4SF18 genes and the sense of transcription (arrows) are indicated. Homology of the syntenic loci was analyzed using the MultiPipMaker alignment building tool. Homologous regions are shown as pale green boxes, and highly conserved regions (more than 100 bp without a gap displaying at least 70% identity) are shown as dark green boxes.