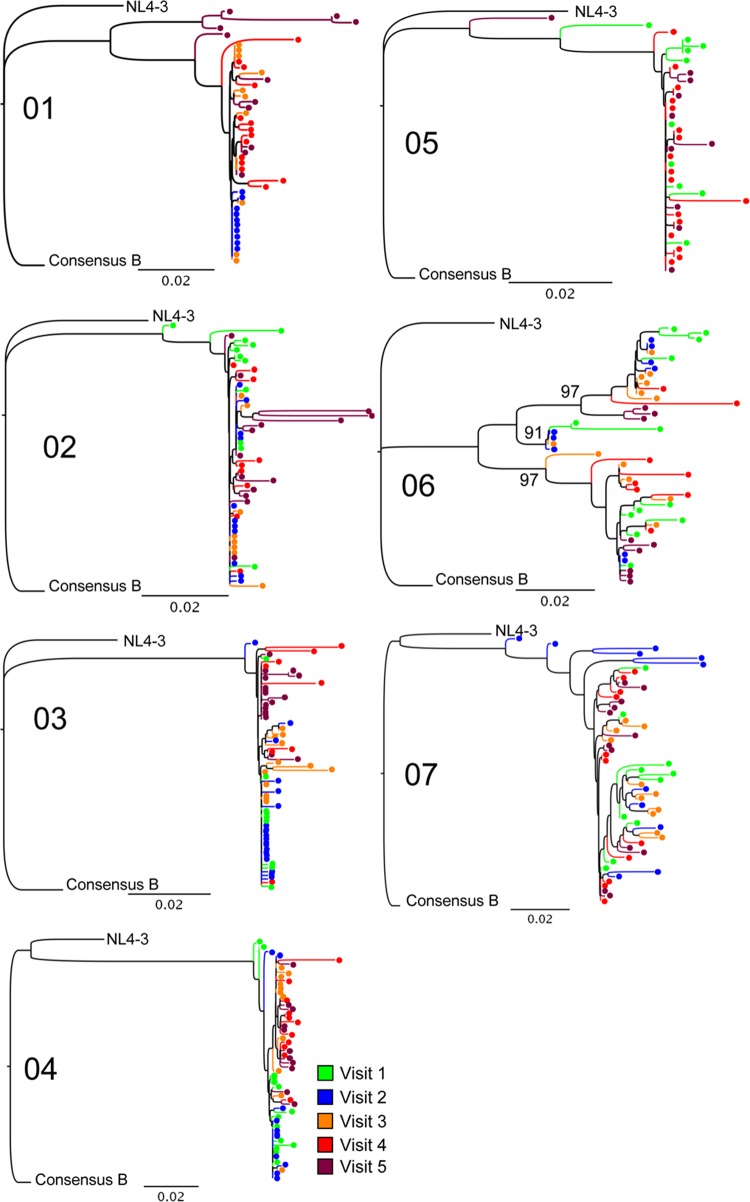
FIG 2.
Phylogenetic trees demonstrate transmitted virus and early diversification. Multiple full-length nef isolates were sequenced at 3 to 5 time points, about 2 weeks apart (Fig. 1). Isolates are color-coded by study visit number as indicated in the key. Sequences were aligned, edited, and translated in comparison to both the NL4-3 and the clade B consensus (“Consensus B”) sequence. Neighbor-joining trees were constructed and evaluated with 1,000 bootstrap replicates. Sequences from each subject formed independent clusters with 100% bootstrap support. As noted, the sequences for subject 07, who was in the early chronic phase of infection, served as a control for comparison. Of the acutely infected subjects, only subject 06 had evidence of multiple transmitted variants, with 3 statistically significant clusters supported by high bootstrap values (91%, 97%, and 97%), as indicated on the tree. Bars, 0.02 genetic distance.