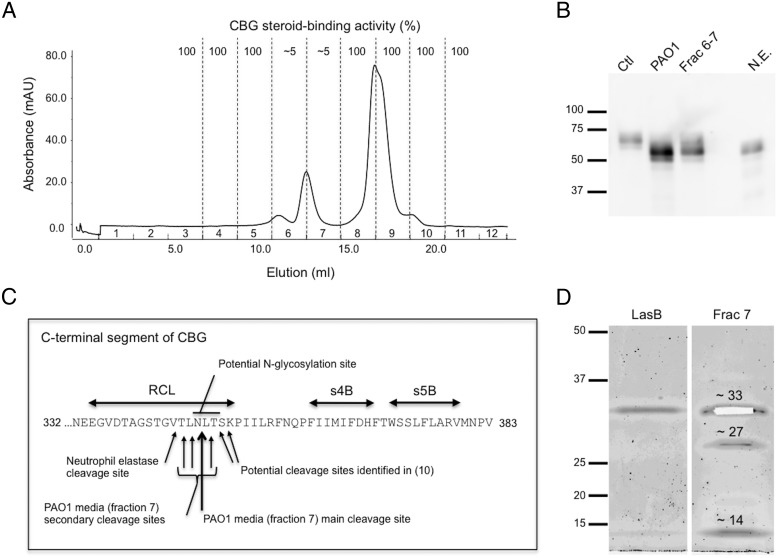
Figure 3.
Chromatographic separation and identification of candidate proteases within P. aeruginosa media that target and cleave the CBG reactive center loop (RCL). A, Elution profile of proteins from a semipurified extract of P. aeruginosa medium after Superdex75 chromatography (see Methods). Fractions 6 and 7 disrupted the cortisol-binding activity of CBG. B, Purified CBG was incubated with P. aeruginosa media (PAO1), with active chromatographic fractions of P. aeruginosa media (Frac 6–7), or with neutrophil elastase (N.E.), and subjected to SDS-PAGE. A shift in molecular weight of 5–10 kDa is observed in all treatment conditions. Protein ladder (kDa) is shown on the left. C, The RCL and adjacent β-sheets (s4B and s5B) are indicated within the C-terminal region of human CBG. Mass spectrometry was performed after incubation of human CBG with fraction 7 (panel A). The main cleavage site was located between N347 and L348 (large arrow). The consensus sequence for N-glycosylation, as well as the neutrophil elastase cleavage site (5, 6) and the cleavage sites observed previously in a human CBG crystal structure (10) are indicated. D, Sypro Ruby stained SDS-PAGE of fraction 7 revealed the presence of 3 bands that were excised for analysis by mass spectrometry (see Methods and Supplemental Data). The approximately 33 kDa band (excised from the gel) was identified as LasB, the approximately 27 kDa band as protease IV, and the approximately 14 kDa band as a C-terminal fragment of LasB. LasB obtained commercially is also shown as a reference.