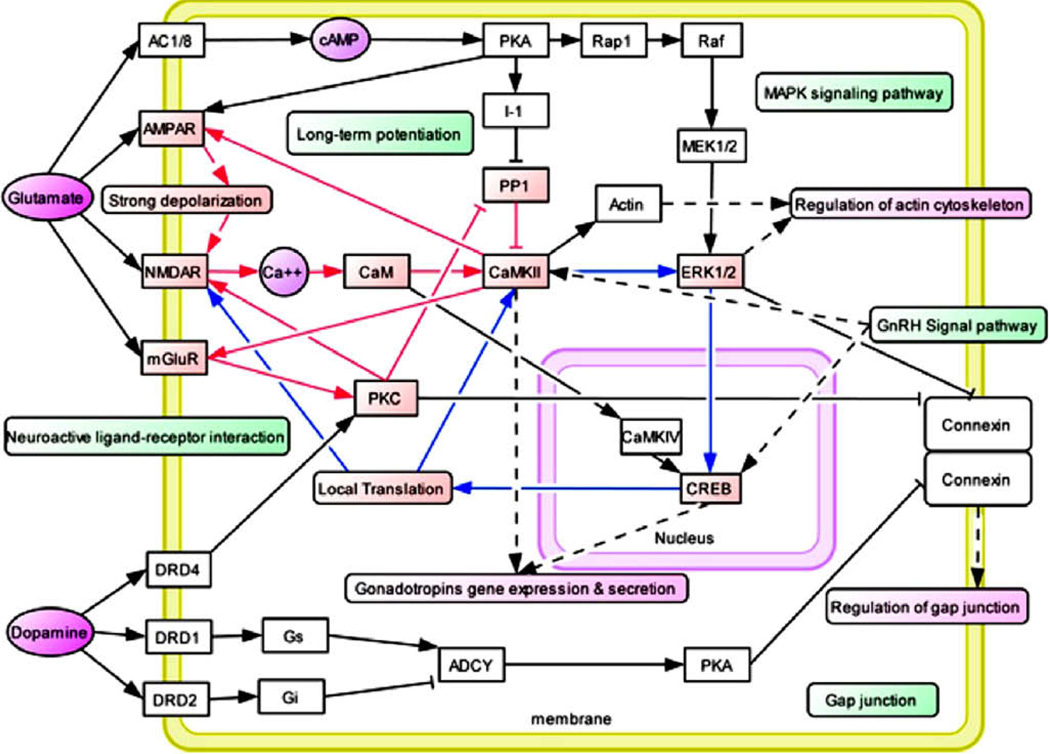
Figure 2.
Hypothetical Common Molecular Network for Drug Addiction (Li et al., 2008 with permission). Most recently Li and his associates [41] developed an addiction gene network that was constructed manually based on the common pathways identified in their 2008 study and protein interaction data. Addiction-related genes were represented as white boxes, while neurotransmitters and secondary massagers were highlighted in purple [29]. The common pathways are highlighted in green boxes. Related functional modules such as “regulation of cytoskeleton”, “regulation of cell cycle”, “regulation of gap junction”, and “gene expression and secretion of gonadotropins” were highlighted in carmine boxes. Several positive feedback loops were identified in this network. Fast positive feedback loops were highlighted in red lines, and slow ones were highlighted in blue lines.