Figure 2. HSV replication fork and model for model for coordinated leading- and lagging-strand DNA synthesis.
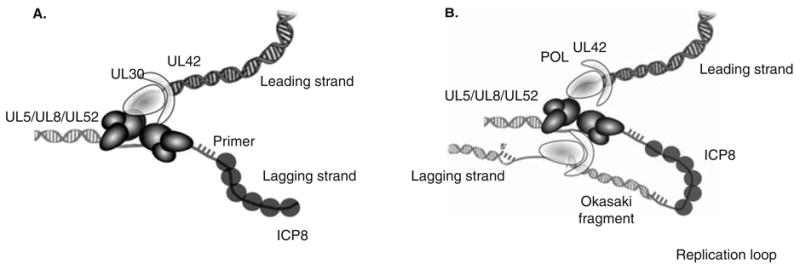
A. An HSV-1 replication fork would be expected to contain the helicase–primase complex (UL5/UL52/UL8) at the fork: UL5 would be expected to unwind duplex DNA ahead of the fork, and UL52 would be expected to lay down RNA primers that the two-subunit DNA polymerase (UL30/UL42) extends. The HSV-1 Pol also carries out leading-strand synthesis. ICP8 (UL29, SSB) presumably binds to ssDNA generated during HSV DNA synthesis. B. Coupled leading- and lagging-strand synthesis was recently reconstituted in vitro and requires HSV polymerase, helicase–primase and ICP8 [74]. In this model, coupled leading-and lagging-strand DNA synthesis requires two heterotrimers of helicase/primase and two molecules of the DNA polymerase. The leading- and lagging-strand polymerase molecules are proposed to communicate with each other through interactions between UL30 and the helicase–primase. A replication loop is formed in the lagging-strand to align it with the leading-strand. The lagging-strand DNA polymerase initiates Okazaki fragment synthesis using RNA primers (hatched bars).
