Fig. 6. Altered histone modifications and transcription factor recruitment at the Ebf1 promoter locus in Mysm1-/-B-cell progenitors.
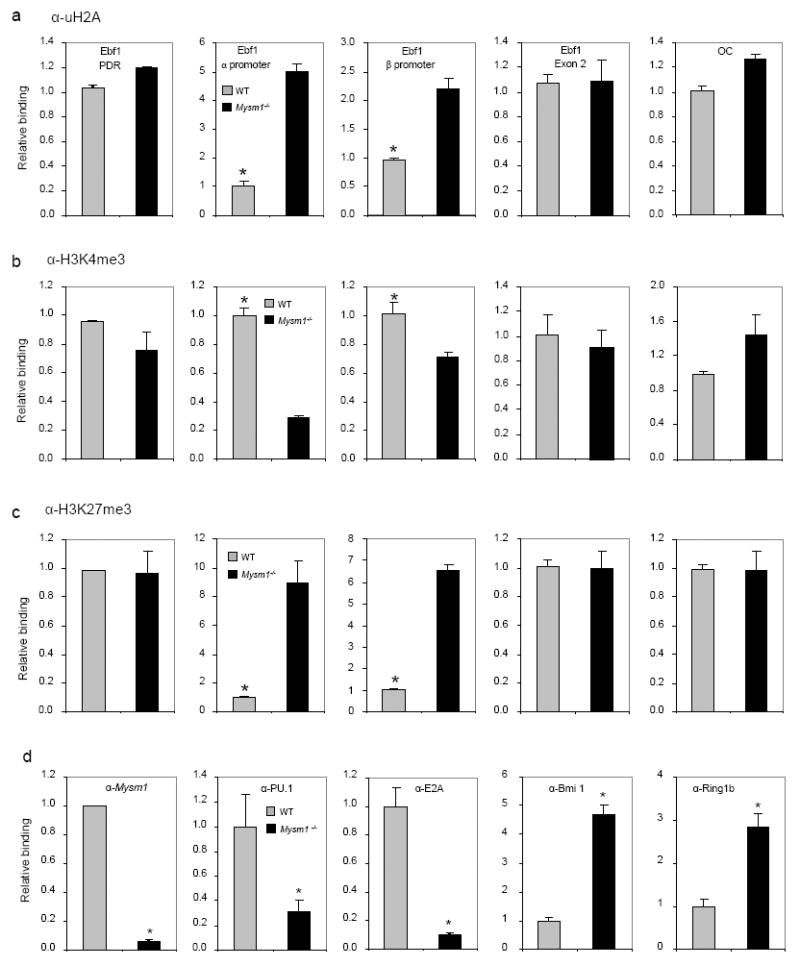
a-c. ChIP analyses of WT or Mysm1-/-BM Lin− cells using uH2A (a), H3K4me3 (b), H3K27me3 antibody (c), or IgG control. The precipitated DNA was analyzed by quantitative PCR using primers amplifying the EBF1α or EBF1β promoter regions, the EBF1 PDR region, the EBF1 exon 2 region, or the control OC promoter region, and normalized with the input DNA before compared to WT (set as 1). *p < 0.05, WT vs. Mysm1-/-.
d. ChIP analyses of the occupancy of E2A, PU.1, Bmi1 and Ring1B in WT or Mysm1-/-BM Lin− cells using a panel of antibodies indicated. The precipitated DNA was analyzed by quantitative PCR using primers (primer pair A) amplifying the EBF1α promoter region for ChIP with antibodies against E2A, Bmi1, Ring1B and MYSM1 and using primers (primer pair E) amplifying the EBF1β promoter region for ChIP with anti-PU.1, and normalized with the input DNA before compared to WT (set as 1). *p < 0.05.
