Figure 1. C9ORF72 ALS iPSNs Exhibit GGGGCCexp Repeat Allele and Reduced C9ORF72.
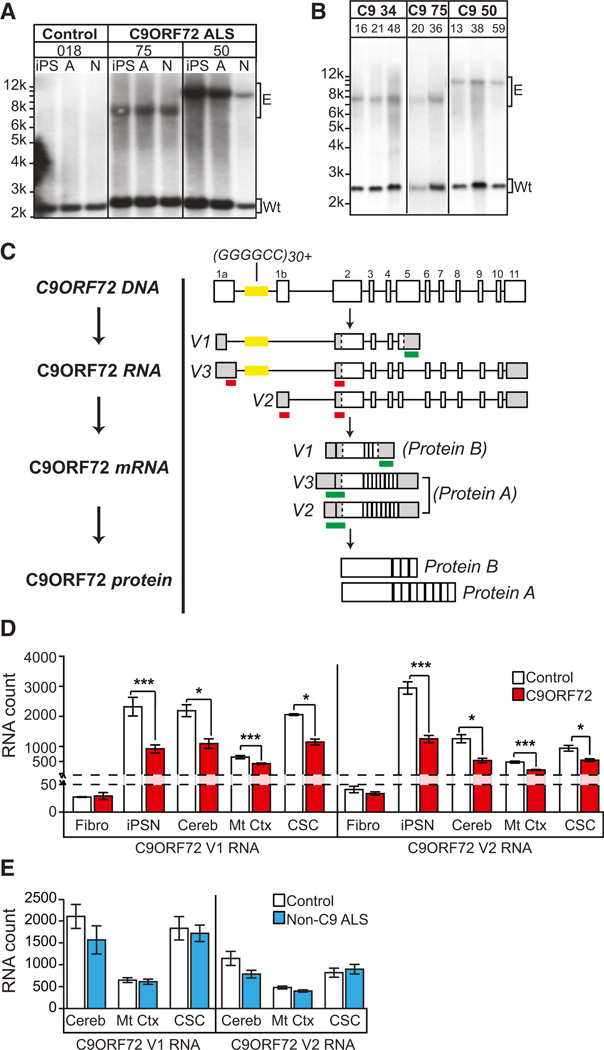
(A) Southern blot of C9ORF72 ALS fibroblasts and subsequent iPSCs for lines used in this study. All C9ORF72 lines contained normal length allele (Wt) and an expanded ‘‘GGGGCC’’ repeat allele (E) that is maintained during differentiation to iPSN (N) or astrocytes (A).
(B) Repeat length of iPSCs was maintained over several iPSC cell passage numbers (ranging from p13 to p59).
(C) Schematic of the C9ORF72 gene, location of the repeat sequence (yellow), validated RNA variants, and location of the nanostring probesets. For detection of nanostring probe, each individual probe (red) must bind in tandem for detection (green). Probes to C9ORF72 V1 will detect pre-mRNA and mRNA; probes to V2 and V3 will only detect C9ORF72 mRNA.
(D) C9ORF72 ALS iPSNs and CNS tissue exhibit reduced C9ORF72 RNA levels (n for C9ORF72 versus Control: n = (fibroblast, Fibro) 5 versus 5; (iPSN) 3 versus 3; (cerebellum, Cereb) 4 versus 6; (motor cortex, Mt Ctx) 10 versus 6; (cervical spinal cord, CSC) 4 versus 6).
(E) Non-C9ORF72 ALS patients do not show attenuated levels of C9ORF72 RNA.
(*p < 0.05; ***p < 0.001). See also Figures S1 and S2; Tables S1, S2, and S3.