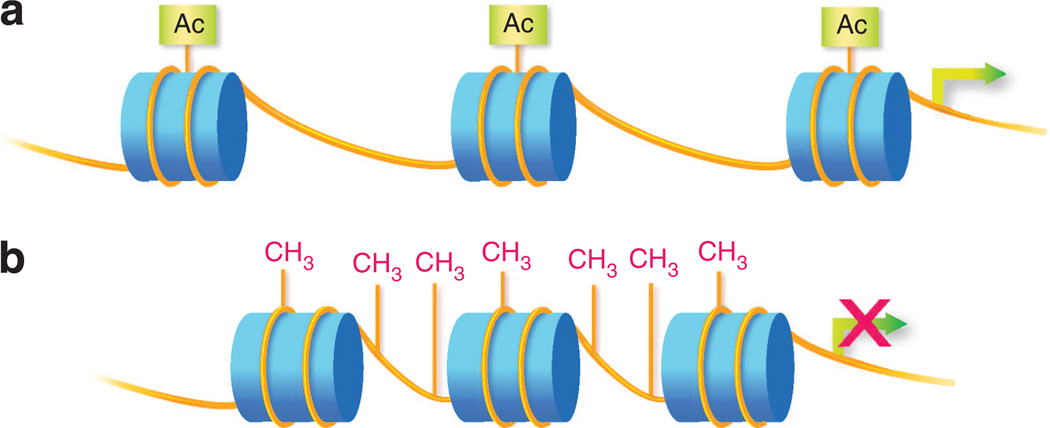
Figure 1. Epigenetic modifications.
(a) Histone modification. Histone proteins (depicted as blue cylinders) spool DNA (depicted as yellow lines) to form nucleosomes and chromatin. Posttranslational modifications (e.g., addition of acetyl, methyl, or ubiqituin groups) to the N-terminal tail regions of histones alter local chromatin conformation. Variability in chromatin condensation affects the accessibility of genes. Loosely condensed regions (euchromatin) are more actively expressed and tightly condensed regions (heterochromatin) are repressed. Depicted in the figure is histone acetylation, associated with loosening of local chromatin and more active gene expression. (b) DNA methylation. DNA methylation occurs through the addition of a methyl group to the C5 position of cytosine to form 5-methylcytosine, typically at cytosine phosphate guanine (CpG) dinucleotides. In promoter regions, DNA methylation silences genes by interfering with transcription factor binding and/or recruitment of methyl-CpG-binding proteins that recruit repressor complexes. DNA methylation is typically associated with tightly condensed chromatin.