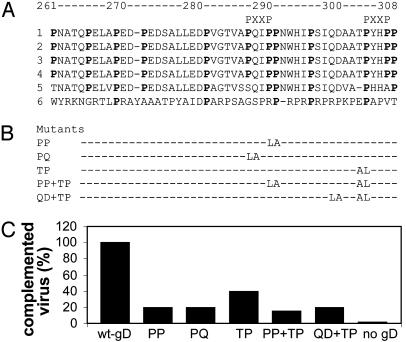
Fig. 7.
(A) Alignment of gD sequences from HSV-1, strains F (1), KOS (2), Patton (3), ANG (4), HSV-2 strain HG52 (5), and pseudorabies virus strain Kaplan (6). Proline residues are bold. In pseudorabies virus, conserved prolines are bold. Minimal motifs (PXXP) able to bind SH3 domains are marked. (B) Designation of proline mutants and engineered substitutions. (C) Infectivity complementation of gD-/+ HSV by gD proline mutants. Transfections, infections, and virion titrations were as detailed in Fig. 5C.