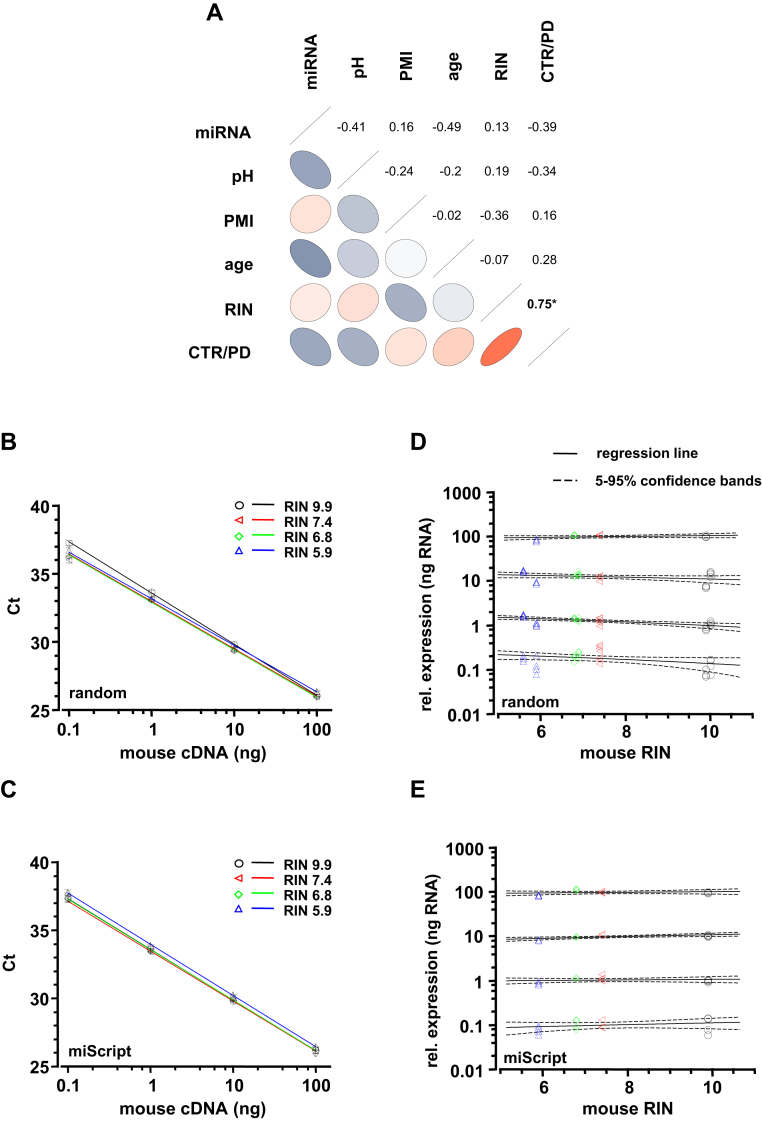
Fig. 1.
Characterization of human postmortem brain samples and evaluation of RT-qPCR protocols for quantification of mRNA and miRNA. (A) Partial correlations between given parameters of brains analyzed in this study. Correlations between 2 parameters were controlled by the other 4 parameters in each calculation. Positive (red) or negative (blue) correlation given by orientation and color of ellipses, strength of correlation given by color intensity, and shape of ellipses, significant correlation indicated by asterisks. Note the strong significant correlation between RNA quality (given as RNA integrity number; RIN) and disease state (CTR vs. PD), due to significant difference of RIN values between control (CTR, n = 8) and PD (n = 5) brains (see also Table 1). Other correlations were not significant. (B–E) Sensitivity and reproducibility of both used RT-qPCR protocols (B/D: random primer protocol and C/E: miScript OligodT primer protocol) was independent of integrity-levels of RNA used as templates for cDNA synthesis (RIN range: 9.9–5.7 of mouse cDNA, covering the RIN-spectrum of the human samples of this study). (B/C) Note that sensitivity of qPCR assay (mLdh-2) is similar for all midbrain cDNA samples, each with 4 different RIN as templates (p > 0.1 in all comparisons; samples generated by thermal degradation of the same midbrain derived RNA for 0–72 minutes at 70 °C). (D/E) Results for RIN 9.9 from panels B and C (standard curve) were used for calculation of relative expression levels of mLdh-2 at different RIN values (range: 9.9–5.9). Regression lines with confidence bands show no significant dependence of gene expression levels from RIN values at all dilutions. Abbreviations: cDNA, complementary DNA; CTR, control; mRNA, messenger RNA; miRNA, microRNA; PD, Parkinson's disease; PMI, postmortem interval; RT-qPCR, quantitative real-time polymerase chain reaction.