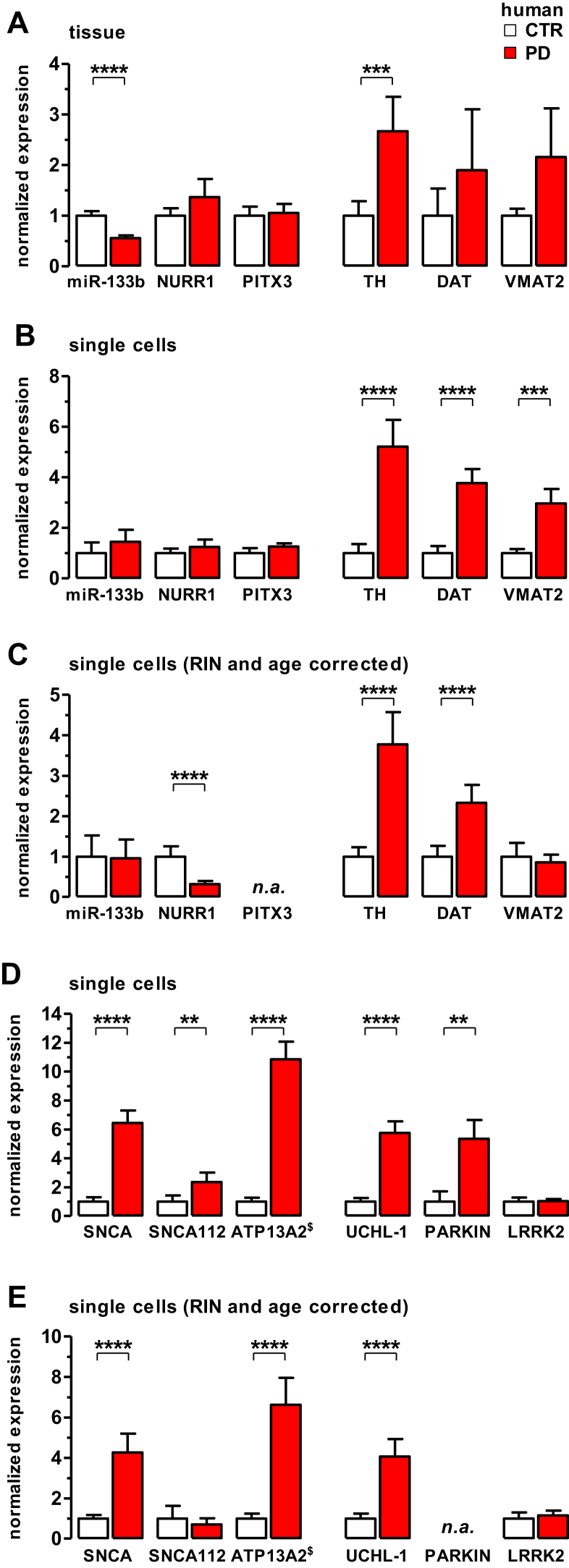
Fig. 3.
Elevated mRNA levels of dopamine release genes and PARK genes in SNCA-overexpressing SN DA neurons in sporadic PD patients. (A) Levels of miR-133b and mRNAs for genes involved in DA neuron development and/or maintenance (NURR1, PITX3) and dopamine homeostasis (TH, DAT, VMAT2), determined via RT-qPCR at the level of midbrain tissue. Relative values, normalized to a geometrical mean of β-actin, ENO2, and TIF-1A. Note significantly lower expression of miR-133b and significantly higher levels of only TH in PD SN tissue compared with controls. (B–C) RT-qPCR analysis of genes as in (A), but at the cell-specific level of individual SN DA neurons, without (B) and after (C) adjustment of data for RIN and age effects. Note that miR-133b levels are not altered in SN DA neurons from PD compared with controls. (D and E) mRNA levels of PARK-genes in individual SN DA neurons, without (D) and after (E) model adjustment of data for RIN and age effects. n.a: model-analysis not possible due to low numbers of samples with positive RT-qPCR results. Bar graphs represent normalized expression as mean ± SEM. For details, see text. Abbreviations: DAT, dopamine transporter; mRNA, messenger RNA; PD, Parkinson's disease; RT-qPCR, quantitative real-time polymerase chain reaction; SEM, standard error of the mean; SN DA, substantia nigra dopamine neurons; TH, tyrosine hydroxylase; VMAT2, vesicular monoamine transporter 2.