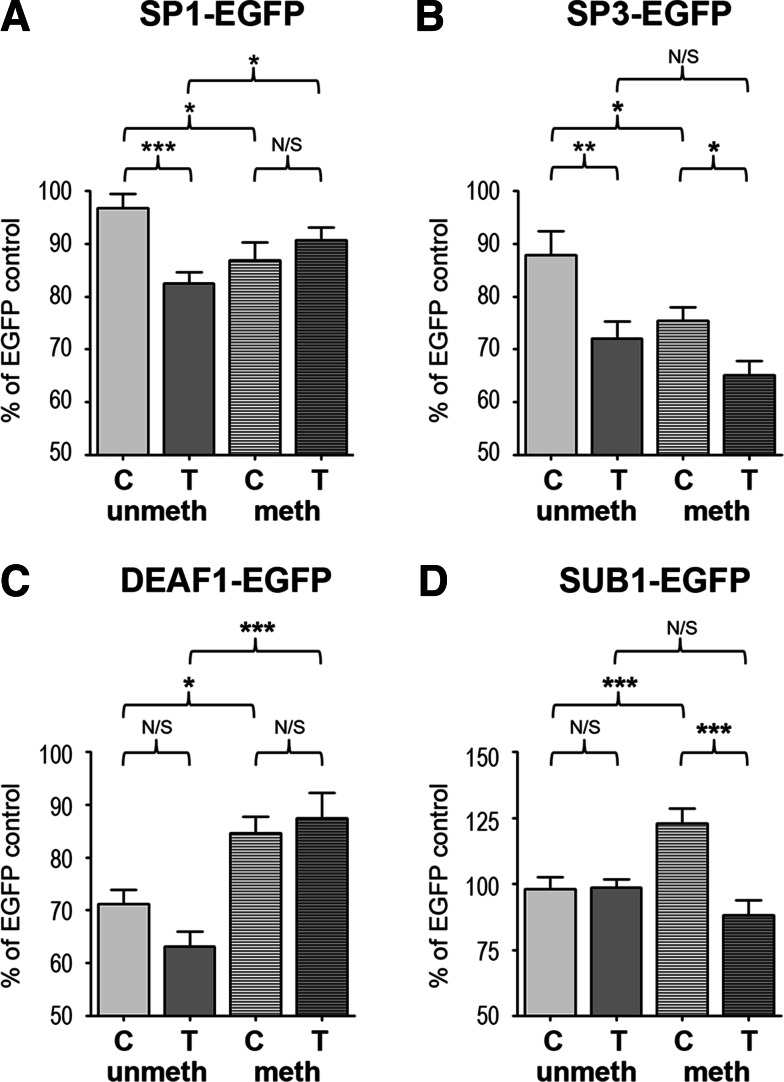
Fig. 6.
Methylation status influences the effect SP1, SP3, SUB1 and DEAF1 have on promoter activity. Unmethylated and in vitro methylated C luciferase vectors containing the GDF5 C or T allele 5ʹUTR were cotransfected into SW1353 cells with (a) SP1-EGFP, (b) SP3-EGFP, (c) SUB1-EGFP or (d) DEAF1-EGFP plasmids and the control pRL-TK Renilla plasmid. Luciferase values were normalised to Renilla and are plotted as a percentage relative to the control co-transfection of the empty EGFP plasmid with the relevant unmethylated or methylated C or T allele 5ʹUTR vector. Data shown are the mean ± standard error for four independent experiments (n = 5) per cell line. A 2-way ANOVA was performed to test for methylation, differences between the alleles and their interaction. Pairwise comparisons were performed using Fisher’s LSD test to compare the four groups, *p < 0.05, **p < 0.01, ***p < 0.001