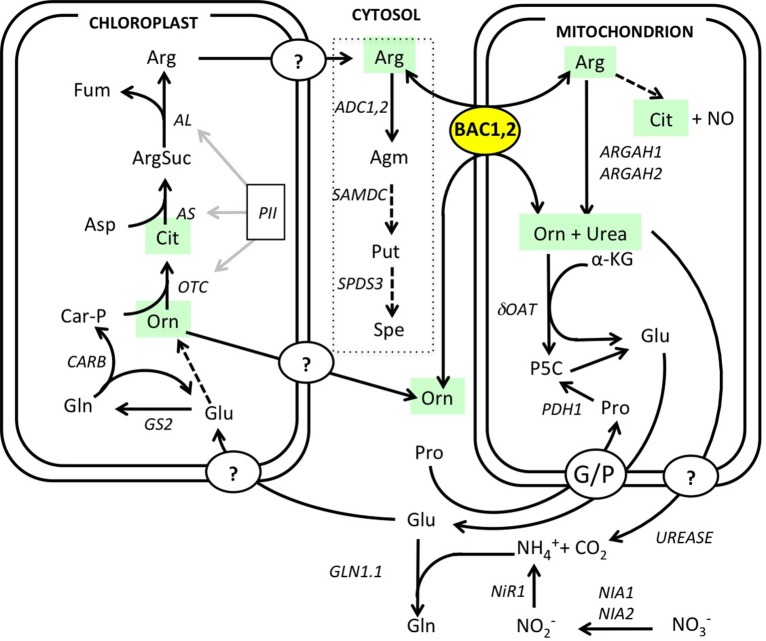
Figure 1.
Overview of arginine mitochondrial transport and metabolism. Arginine metabolism map drawn according to Slocum (2005), Funck et al. (2008), and Witte (2011). Polyamine biosynthesis pathway is outlined (dotted line). Dotted arrows represent pathways with additional steps that are not detailed here. Enzyme and protein abbreviations and references: ADC, arginine decarboxylase (ADC1 At2g16500, ADC2 At4g3471); AL, argininosuccinate lyase (At5g10920); AS, argininosuccinate synthase (At4g24830); ARGAH, arginase (ARGAH1 At4g08900, ARGAH2, At4g08870); BAC, basic amino acid carriers BAC1 (At2g33820) and BAC2 (At1g79900); GS, glutamine synthetase (GS2, At5g35630); GLN, glutamine synthetase (GLN1.1 At5g37600); NIA, nitrate reductase (NIA1 At1g77760, NIA2 At1g37130); NiR1, nitrite reductase (At2g15620); dOAT, delta ornithine amino transferase (At5g46180); OTC, ornithine carbamoyl phosphate transferase (At1g75330); PII, nitrogen sensing protein (At4g01900); PRODH1, proline dehydrogenase (At3g30775); SAMDC, S-adenosine methionine decarboxylase (At3g02470); SPDS, spermidine synthase (SPDS3 At5g53120); urease (At1g67550). Metabolite abbreviations: Agm, agmatine; Arg, arginine; ArgSuc, argininosuccinate; Car-P, carbamoyl phosphate; Cit, citrulline; Fum, fumarate; Gln, glutamine; Glu, glutamate; αKG, alpha-ketoglutarate; NO, nitric oxide; Orn, ornithine; Pro, proline; Put, putrescine; P5C, delta1-pyrroline-5-carboxylate; Spe, spermidine.