Figure 3.
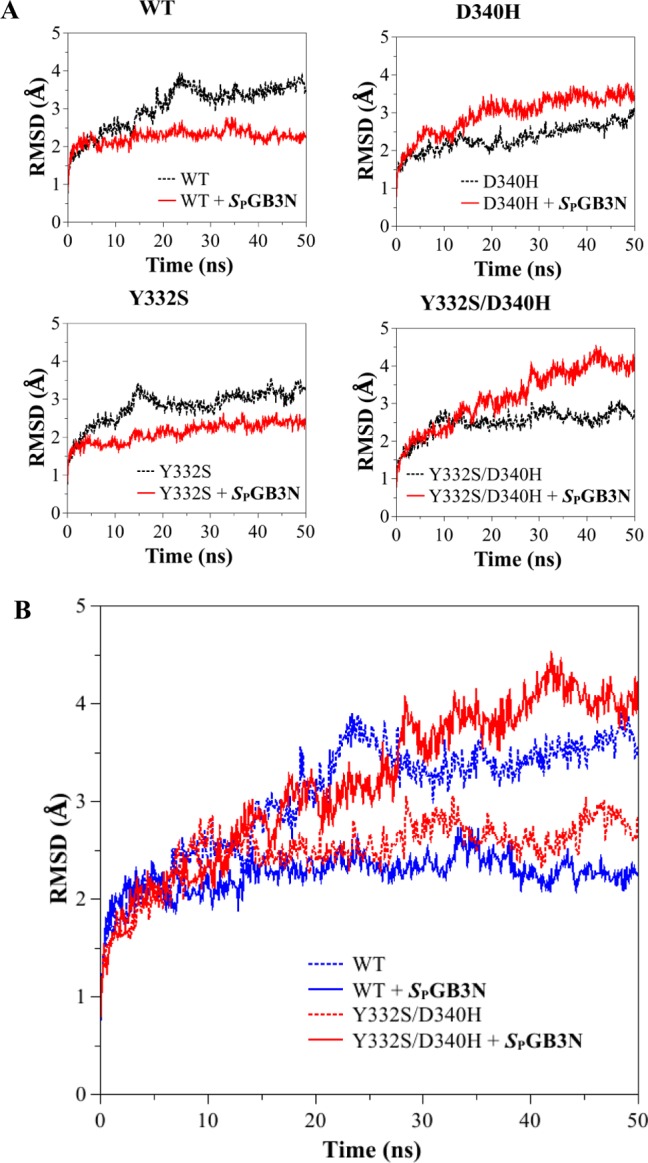
(A) Root-mean-square deviation (rmsd) of the backbone atoms (carbonyl carbon, C; α-carbon, CA; nitrogen, N) from the starting structure (PDB entry 1P0I) as a function of time (nanoseconds), for MD simulations of the WT and hBChE variants. MD simulations were used to compare the starting structure as a function of time for WT and Y332S, D340H, and Y332S/D340H hBChE variants in the absence (black line) and presence (red line) of SPGB3N. (B) Plot of rmsd vs time of all the backbone atoms (C, CA, and N) from the starting structure as a function of time for MD simulations of WT hBChE (blue) and the Y332S/D340H hBChE variant (red) in the presence (solid lines) or absence (dotted lines) of SPGB3N bound to the active site.
