Figure 1.
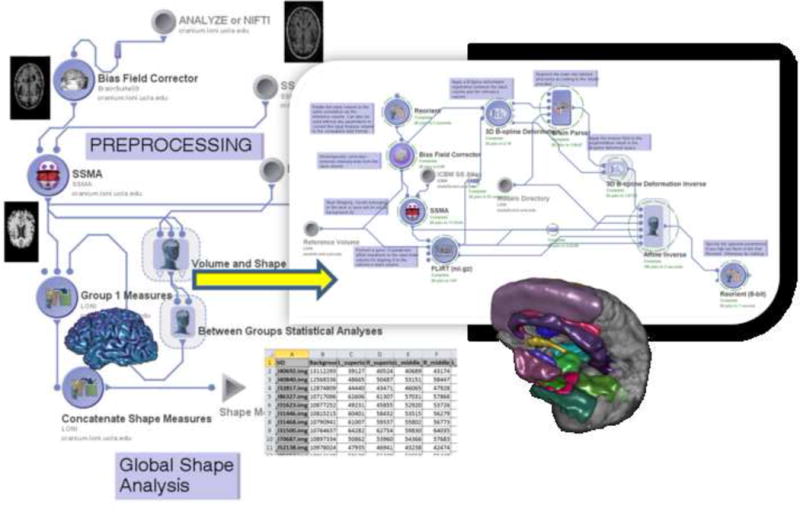
Volumetric pipeline workflow.
This workflow represents a heterogeneous computational protocol that includes data preprocessing (inhomogeneity correction, skull stripping), brain parcellation, volumetric calculation and statistical analysis of derived neuroimaging biomarkers (i.e., volume) and various subject phenotypes (e.g., diagnosis, age). The insert image illustrates part of the nested processing modules part of the volume and shape processing step of the protocol. Examples of outputs of key processing modules are shown as thumbnail images throughout the workflow. The nodes in the pipeline workflow graph represent atomic processing tools or nested groups of data analysis modules. The edges connecting different nodes in the pipeline graph indicate the data flow and the dependencies of the execution protocol. The volumetric workflow pipeline consists of 4 main components (each annotated by a label) – data preprocessing (intensity inhomogeneity correction and skull-stripping, cortical surface modeling, and tissue classification.
Abbreviations : SSMA- skull-stripping meta-algorithm.
