Figure 4. miRNAs DE by cell type.
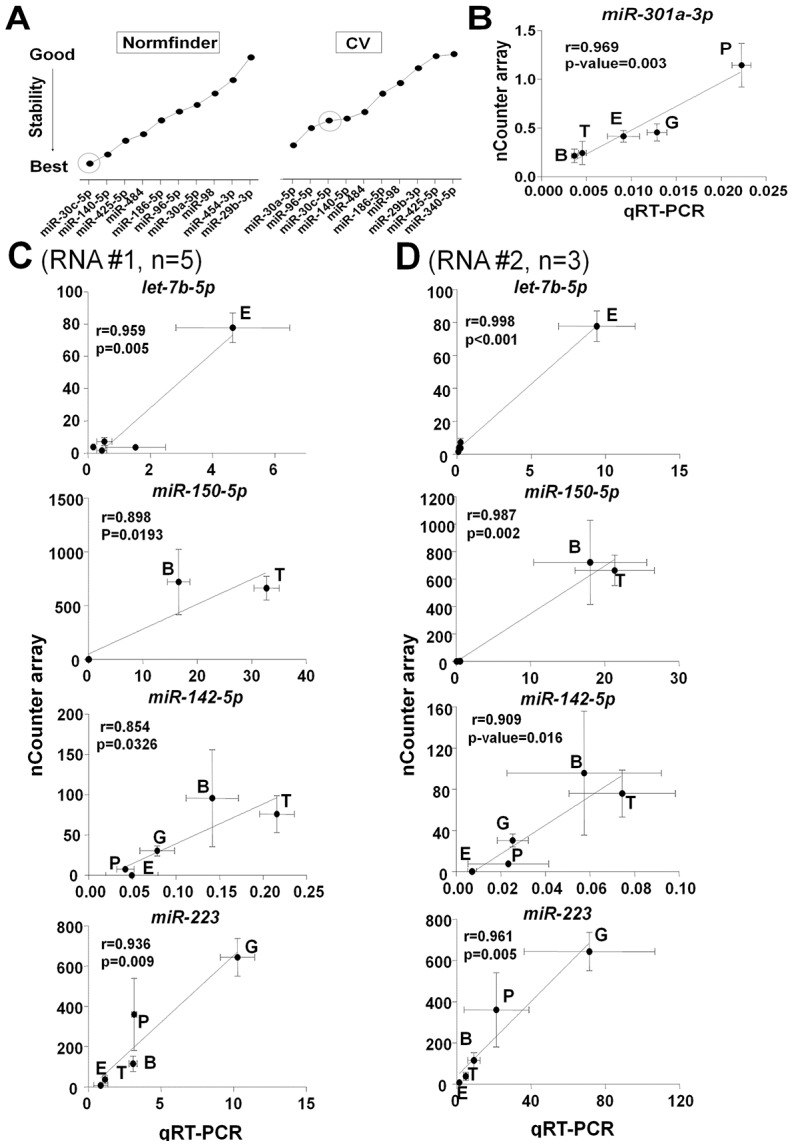
(A) The 10 most stable miRNAs across all cell types are shown for both NormFinder (left) and Coefficient of Variation (CV) methods (right). (B) Validation of NanoString-derived data for miR-301a-3pby qRT-PCR data. The X-axis represents the expression level of each miRNA normalized to miR-30c-3p using 2−ΔCt method. Each point represents the mean ± SEM of 5 subjects for each cell type. (C,D) Validation of microRNAs DE by cell type. The NanoString-derived data for the indicated miRNAs was validated by qRT-PCR using RNA isolated from the 5 hematopoietic cell types from two different preparations of cells and RNA. qRT-PCR data was normalized and presented as in Figure 4B.P, platelets; T, T-cells; B, B-cells; G, granulocytes; E, erythrocytes.
