Figure 1.
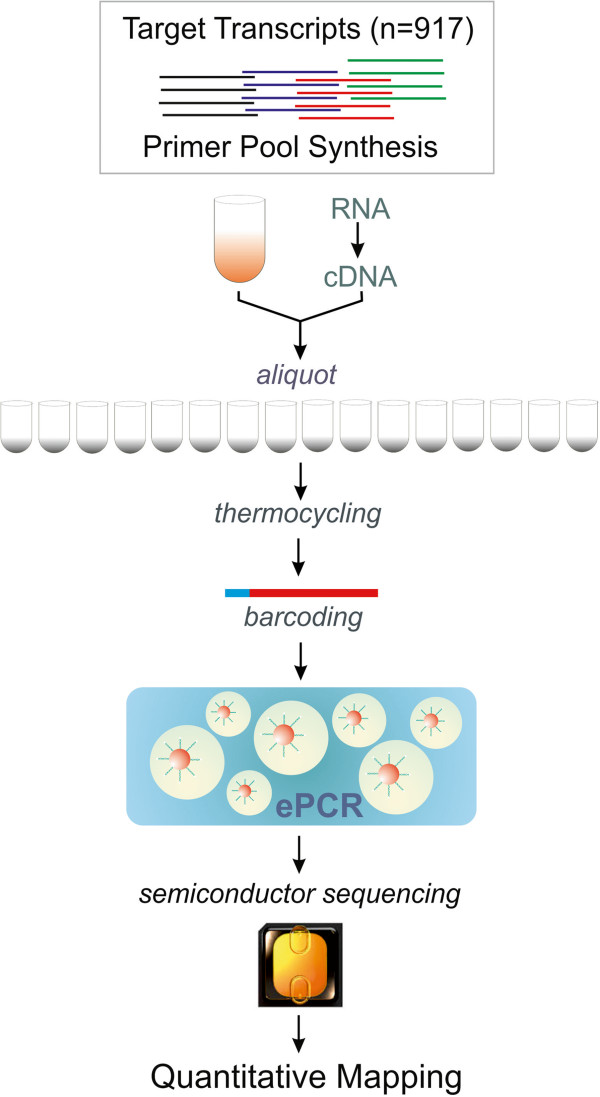
Experimental workflow for AmpliSeq-RNA based transcript quantification. An aliquot of the custom primer pool, the cDNA sample and a PCR master-cocktail are combined in individual tubes or multititer plate wells. Following pre-amplification the amplicons are tagged with a sample specific molecular barcode followed by pooling and emulsion PCR (ePCR) amplification on nanosphere-beads. After semiconductor sequencing (Ion-Torrent-Proton) reads are mapped to the target sequence library. Read frequencies proportional to transcript abundance are provided in a standard spread-sheet for further analysis (for experimental details see Methods Section).
