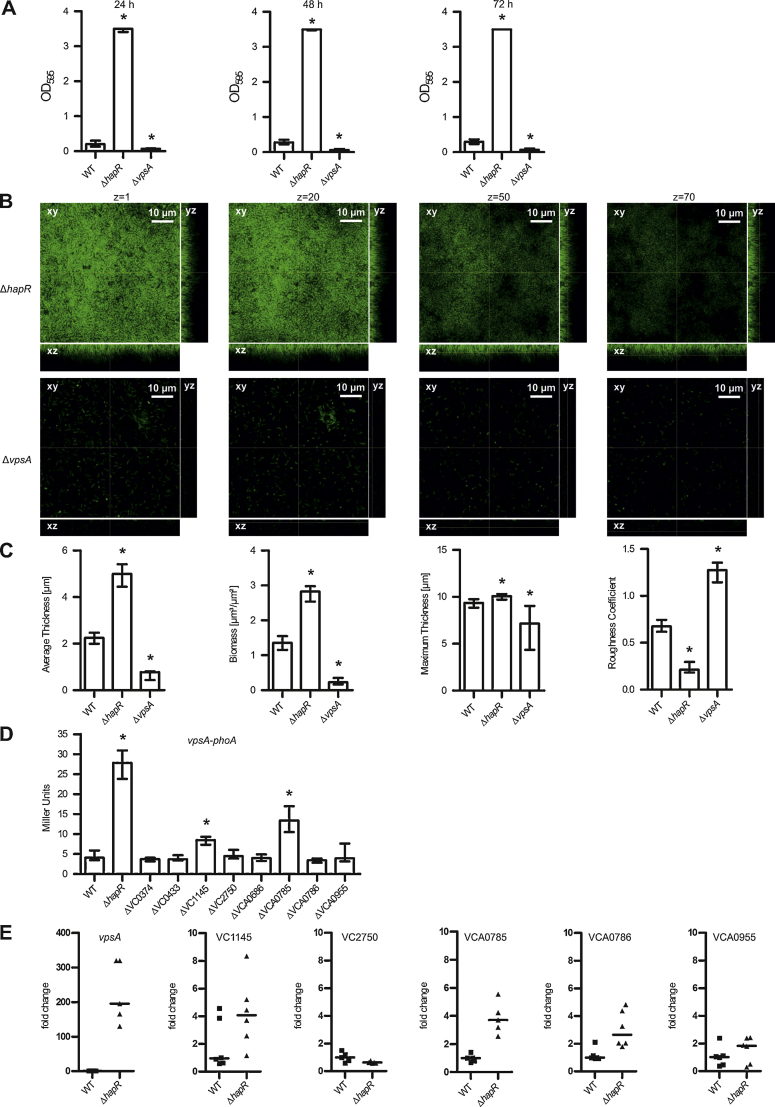
Fig. 6.
Interplay between vps gene regulation, HapR regulation and ibi genes. (A) The biofilm formation capacity of wild type (WT) and deletion mutants was quantified by crystal violet staining and subsequent determination of the OD595 after 24 h, 48 h and 72 h under static conditions. (B) Shown are confocal laser scanning microscopy images of GFP expressing ΔhapR and ΔvpsA mutant biofilms as horizontal (xy) and vertical (xz and yz) projections (large and side panels, respectively). Large panels represent selected single optical sections through the acquired three-dimensional data sets at the indicated z position. Biofilms were grown for 24 h in flow cell chambers with constant 2% LB medium flow. Images of biofilms were acquired using a Leica SP5 confocal microscope with spectral detection and a Leica HCX PL APO CS 63x water objective (NA 1.2). (C) Shown are the respective structural parameters of image stacks of WT and deletion mutants analyzed using the COMSTAT software (http://www.comstat.dk) (Heydorn et al., 2000; Vorregaard et al.). (D) Alkaline phosphatase activities (in Miller Units) were measured from 8 h statically grown cultures of indicated strains harboring a chromosomal vpsA–phoA transcriptional fusion. (A–D) Shown are the medians from at least six independent measurements. The error bars indicate the interquartile range. Significant differences between the respective data sets of the WT compared to mutants (*P < 0.05 Kruskal–Wallis test followed by post hoc Dunn's multiple comparisons) are indicated. (E) Bacterial RNA was extracted from cultures grown statically for 24 h, reverse transcribed to cDNA and used as template for qRT-PCR analysis of the indicated genes. For each sample, the mean cycle threshold of the test transcript was normalized to the housekeeping gene 16S rRNA and to one randomly selected WT reference sample. The data are presented as medians of six independently grown samples. Significant differences compared to WT were detected for the genes vpsA, VCA0785 and VCA0786 (P < 0.05 Mann–Whitney U test).