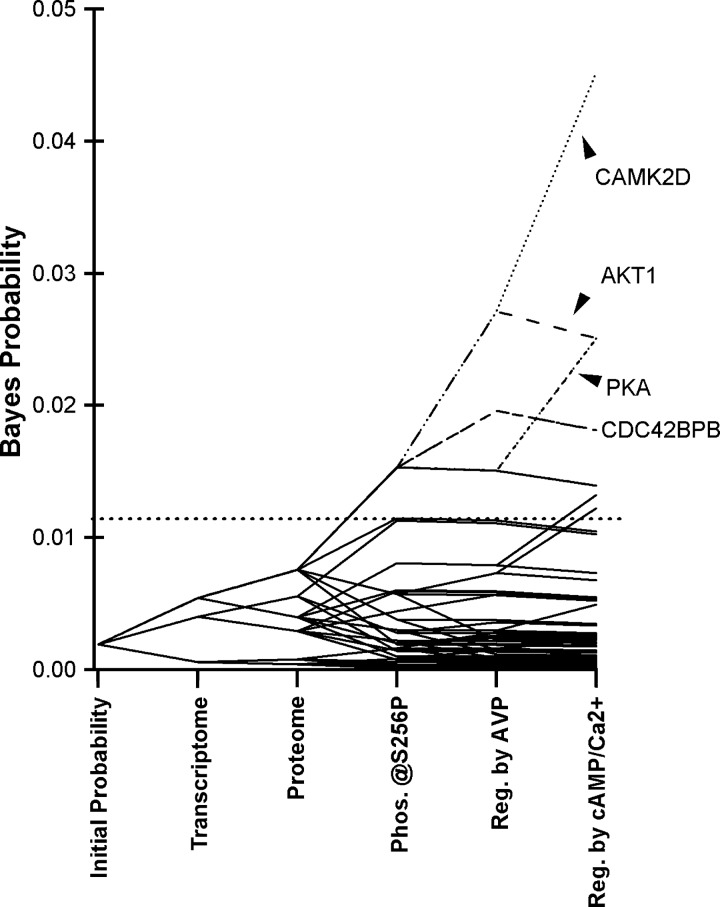
Fig. 3.
Bayes' analysis of rat IMCD kinases. All 521 kinases in the rat genome are assumed to be equally probable at the start of calculations. Probabilities were calculated using Bayes' rule from sequential integration of information from several data sets. Resulting values derived from preexisting information provide “prior probabilities” for assessment of new experimental data presented here. Horizontal dotted line indicates cutoff values for the top 20 kinases. CAMK2D, Ca2+/calmodulin kinase 2δ.