Figure 2.
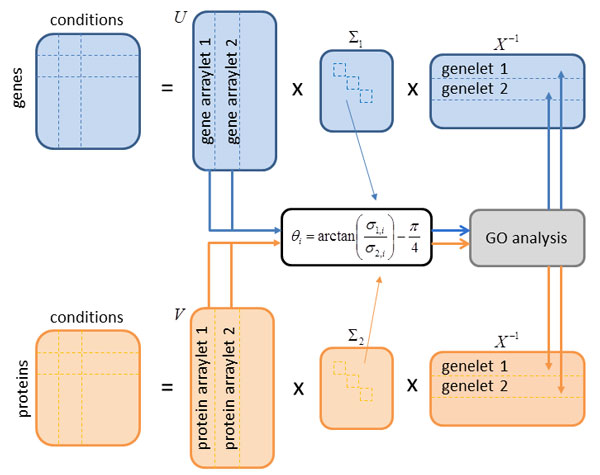
Flowchart of GSVD. The gene expression matrix (blue) contains the genes in the rows and the conditions in the columns. The protein expression matrix (orange) contains the proteins in the rows and the conditions in the columns. The conditions in our study correspond to the life cycle stages of P. falciparum. Genes and proteins annotated to the considered GO terms are gathered in a separate matrix (gray). The gene and the protein expression matrices are each decomposed in three matrices according to equations (1) and (2). The matrices U and V contain the arraylets, which encode for the expression of genes and proteins in the corresponding genelets X-1, which represent the cellular state in the measurement conditions. According to the angular distance θi, which is computed from the generalized eigenvalues σ1,i and σ2,i , a restricted GSE analysis is performed on the genes and/or proteins with the absolute (from a mathematical point of view) highest values in the arraylets in order to assign GO terms to the corresponding genelets. Figure adapted from [3].
