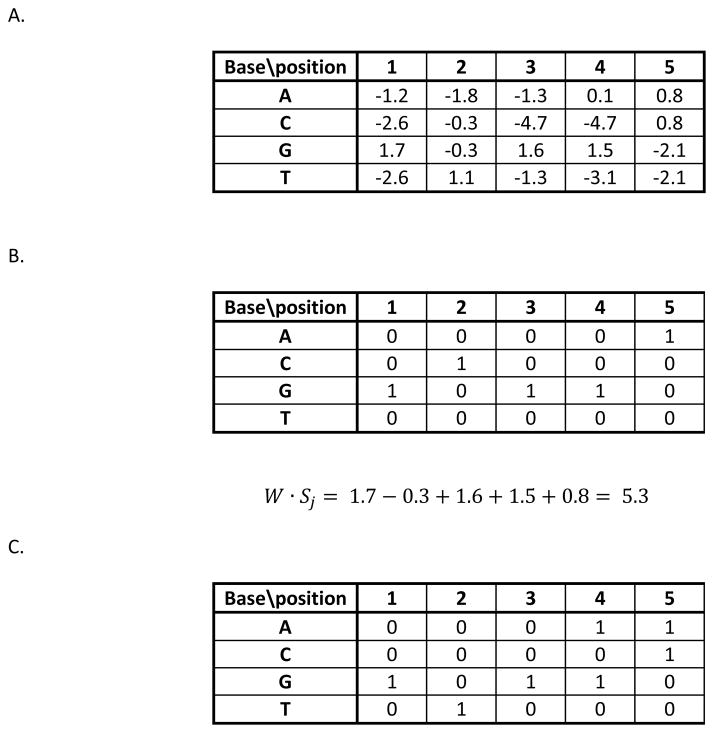
Figure 1.
Weight matrices and sequence encoding. A. The weight matrix for a hypothetical transcription factor (YFTF). Scores are provided for each possible base at each position in a five-long binding site. B. The encoding of a particular sequence, GCGGA, with a 1 for the base that occurs at each position and all other elements are 0. The score of the sequence, given the matrix in part A, is shown. C. An alternative weight matrix for the consensus sequence GCGRM (R=A or T, M=A or C). Any sequence that matches the consensus will get a score of 5, allowing one mismatch requires a score of at least 4, etc. This shows how any consensus sequence can be converted into an equivalent weight matrix that will return exactly the same set of sites.