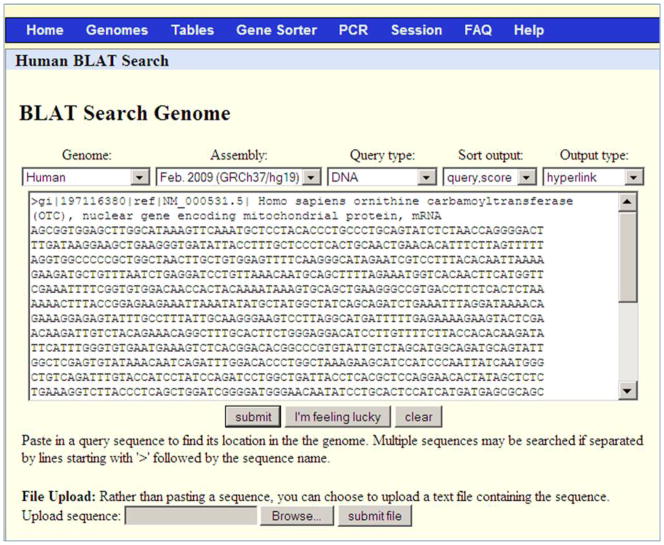
Figure 1.
Web BLAT search screen for Protocol 1. The interface allows the user to easily specify (from left to right) the genome, assembly, mode of search, the desired sorting of the results, and the output format. The Genome pull-down menu provides a choice of over 50 species from mammals, fish, invertebrates, yeast, and others. Some, such as human, will have more than one choice of genome assembly in the Assembly pull-down menu. The Query type pull-down menu provides an ability to choose the mode of the search. The DNA Query type used in this protocol searches a DNA query against a DNA database. Additional available options are described in the text. The Sort output pull-down menu can be used to sort the results table. The options are “query, score”; “query, start”; “chromosome, start”; “chromosome, score”; and “score”. The “query, score” option first sorts by query ID (if multiple sequences are pasted into the input box) and then by score. Finally, the “Output type” pull-down menu provides 3 options to present the results, hyperlink, psl and psl no header. The choice of “Output type” as hyperlink yields a table with a link (Browser) to display each alignment in the UCSC Genome Browser and a link (details) to the details of the alignment. The psl output type provides details about mismatches, gaps, and blocks in a tabular format and does not provide links to alignments or the genome browser. The PSL output format is described in detail in the text. Finally, the large text box is for the input sequence. The sequence needs to be in FASTA format as shown here for the query NM_000531.5, which is used in Protocol 1. Clicking on the submit button generates the table of alignments. The “I’m feeling lucky button” goes directly to the genome browser to display the genome alignment of the best scoring alignment of the first input sequence. The clear button resets the input text box. The “Browse” and “submit file” buttons are for uploading sequences from a file instead of copying them into the text box.
In this Protocol, the selected options are “Human” genome, “Feb 2009 GRCh37/hg19” assembly, “DNA” as query type, “query, score” for sorting the results and “hyperlink” as the output type.