Figure 2.
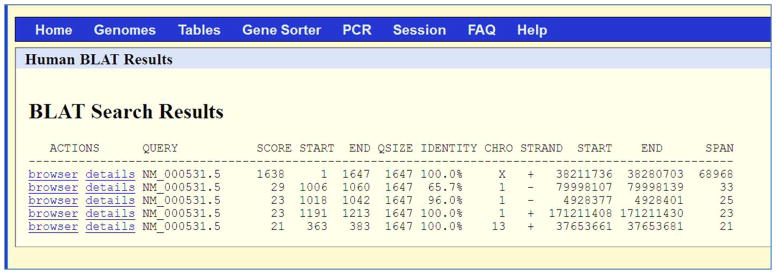
Results table for Protocol 1. The BLAT search results provide the following columns: 1) ACTIONS – links for visualization of the alignment in the UCSC Genome Browser (browser link) and a more detailed alignment text view (details link); 2) QUERY – an identifier for the query sequence; 3) SCORE – the number of matches with a penalty for mismatches and gaps (see subsection “Score calculation” in the Commentary); 4) START – the location of the beginning of the alignment in the query sequence; 5) END – the location of the end of the alignment in the query sequence; 6) QSIZE – the length of the query sequence; 7) IDENTITY – an indication of the number of matching bases and gaps (see subsection “Percent identity calculation” in the Commentary); 8) CHRO – the chromosome; 9) STRAND – both query strands (‘+’ and ‘−’) are checked in the alignment. In the translated alignment mode, a second ‘+’ or ‘−’ for the genomic strand is provided; 10) START – the location of the beginning of the alignment in the genome sequence; 11) END - the location of the end of the alignment in the genome sequence; and 11) SPAN – the number of bases on the genome covered by the alignment. The information in the table – such as score, span and identity – indicates the extent of the match.
The results in this Protocol are sorted by score; the top result has a much higher score than the others. The first row in the results shows that the QUERY NM_000531.5 matches the human genome with a score of 1638 (SCORE column) from its nucleotide 1 to 1647 (START and END columns next to the SCORE column). The query size is 1647 (QSIZE column). Thus, the entire query has coverage in the human genome with 100% identity (IDENTITY column). This alignment is on chromosome X (CHRO column), on the plus/forward strand (STRAND column) from nucleotide 38211736 to 38280703 (START and END columns next to the STRAND column), covering a range of 68968 (SPAN column) nucleotides.
