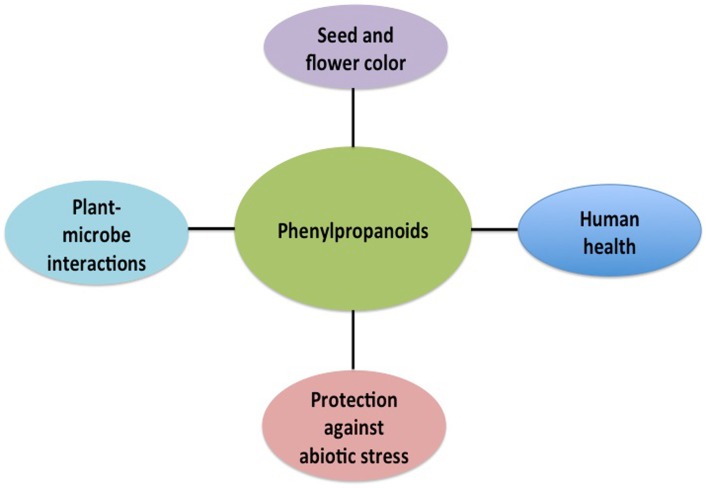
Like other organisms, plants have to deal with a dynamic environment. To respond and adapt to the environmental conditions plants rely in a diverse battery of cell-surface protein-receptors and secondary metabolites. Phenylpropanoids are metabolites required for this big task, they regulate a wide range of physiological process, such as pigmentation of flowers and fruits, seed dispersal through attracting pollinators, auxin transport and UV-B protection (Peer and Murphy, 2007; Tanaka et al., 2008; Agati and Tattini, 2010; Vogt, 2010). Likewise, these secondary metabolites are critical to establish symbiotic interactions as well as to fight against pathogens (Dixon et al., 2002; Cooper, 2004). Besides these roles in planta, phenylpropanoids play an important role in the human health. For example, different flavonoids have neuroprotective, anti-inflammatory, analgesic, bacterial and anti-fungicidal activity (Figure 1; Yu and Jez, 2008).
Figure 1.
Phenylpropanoid derivatives control different physiological aspects at planta level and play an important role in the human health.
One of the most exiting examples about the relevance of phenylpropanoid in planta and human health levels is the common bean (Phaseolus vulgaris). In one hand, this legume uses different flavonoids to establish a symbiotic interaction with the N2-fixing soil bacteria collectively known as rhizobia. Through this interaction, common bean obtains nitrogen for its own metabolic requirements and this in turn is delivered to other organisms, including humans. In the other hand, common bean grains, besides being the main source of protein and fiber for human consumption, provide a variety of flavonoids, isoflavonoids and lignans with potential medical properties (Broughton et al., 2003).
Most of our knowledge about the phenylpropanoid synthesis and the genetic control of this pathway come from biochemical and genetic studies on the model plant Arabidopsis thaliana (Vogt, 2010). Despite the ecological and dietary relevance of common bean, our knowledge about the genetic control of the phenylpropanoids in this legume is scarce. To fill-up this gap, Reinprecht et al. (2013) used comparative genomics analyses and traditional PCR-based cloning to identify 46 structural and regulatory genes from the common bean phenylpropanoid pathway. The identified genes would play roles in: general phenylpropanoid pathway (PAL, C4H, and 4CL), lignin/lignan- (CCR, CAD, C3H), flavonoid/anthocyanin- (CHS, CHI, CHR, F3H) or isoflavonoid (IFS, IFR, 7IOMT) biosynthesis. Interestingly, with these experimental approaches, Reinprecht and colleagues were able to identify the transcription factors LIM, KAP-2, HD, WIP, and Myb15, all of them with a potential role in the transcriptional control of the phenylpropanoid pathway. These results provide a significant insight in our knowledge about the genetic control of the phenylpropanoid pathway in common bean. Furthermore, this study provides two additional aspects: (1) a large repertory of candidate genes for reverse genetic analyses that can lead us to understand their role in the biosynthesis of these secondary metabolites, and (2) a variety of marker genes that can be used for plant breeding programs in common bean.
Mapping genes into the genome is a required step for plant breeding programs. For this, it is also important to know the genome sequence of the plant of interest. For several years, the lack of the common bean genome sequence was one of the main limitations to do a deep genetic analysis. Recently, the common bean genome sequence was reported (Schmutz et al., 2014). This together with the known soybean genome sequence (Schmutz et al., 2010) opens the gates to perform detailed comparative analysis. Reinprecht et al. (2013) harnessed the close genetic relatedness between common bean and soybean to performed a comparative mapping in silico and were able to map the 46 identified-phenylpropanoid genes into some of the 11 chromosomes of common bean. For example, they mapped CHS, Myb4, PAL3, and LIM in the common bean chromosome Pv2, Pv6, Pv8, Pv9, respectively. Perhaps one of the most interesting results of this mapping analysis is the fact that most of the identified transcription factor genes were mapped in the chromosome Pv10. This finding is relevant considering that there are several cases where most of the genes that control a particular pathway are located in one chromosome. For example, several genes that control the symbiotic interaction between Medicago truncatula and rhizobia are located in the chromosome 5 (Ané et al., 2002; Arrighi et al., 2006; Horváth et al., 2011).
Knowing the structural and regulatory genes of the phenylpropanoids pathway will certainly help to understand how these secondary metabolites regulate different physiological processes. Likewise, having their genomic location will contribute to develop breeding programs in common bean. However, it is important to realize that several aspects have to be improved before carrying this out. Perhaps the two most important aspects are: (1) increase the number of marker genes and saturate the common bean genome; (2) develop powerful common bean mapping populations, for instance Nested Association Mapping lines. Indeed, it is widely documented that this kind of mapping population provides a high resolution in mapping analysis (Poland et al., 2011). If we consider that: (1) common bean has a vast genetic diversity (even greater than soybean diversity) and (2) Next Generation Sequencing technology has decreased the cost to get the whole genome sequence of any organism; these two aspect are feasible with no problem. Having a high resolution genetic map and a powerful mapping population will help us to accurately map these genes involved in the phenylpropanoid biosynthesis as well as identify additional genes that might control this pathway.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Agati G., Tattini M. (2010). Multiple functional roles of flavonoids in photoprotection. New Phytol. 186, 786–793 10.1111/j.1469-8137.2010.03269.x [DOI] [PubMed] [Google Scholar]
- Ané J. M., Lévy J., Thoquet P., Kulikova O., de Billy F., Penmetsa V., et al. (2002). Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in nod factor transduction, nodulation, and mycorrhization. Mol. Plant Microbe Interact. 15, 1108–1118 10.1094/MPMI.2002.15.11.1108 [DOI] [PubMed] [Google Scholar]
- Arrighi J. F., Barre A., Ben Amor B., Bersoult A., Soriano L. C., Mirabella R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279 10.1104/pp.106.084657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W. J., Hernandez G., Blair M., Beebe S., Gepts P., Vanderleyden J. (2003). Beans (Phaseolus spp.)—model food legumes. Plant Soil 252, 55–128 10.1023/A:102414671061118055156 [DOI] [Google Scholar]
- Cooper J. E. (2004). Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 41, 1–62 10.1016/S0065-2296(04)41001-5 [DOI] [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C. J., Reddy M. S. S., Wang L. (2002). The phenylpropanoid pathway and plant defence a genomics perspective. Mol. Plant Pathol. 3, 371–390 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- Horváth B., Yeun L. H., Domonkos A., Halász G., Gobbato E., Ayaydin F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24, 1345–1358 10.1094/MPMI-01-11-0015 [DOI] [PubMed] [Google Scholar]
- Peer W. A., Murphy A. S. (2007). Flavonoids and auxin transport: modulator or regulators?. Trends Plant Sci. 12, 556–563 10.1016/j.tplants.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Poland J. E., Bradbury P. J., Bucler E. S., Nelson R. J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. U.S.A. 108, 6893–6898 10.1073/pnas.1010894108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinprecht Y., Yadegari Z., Perry G. E., Siddiqua M., Wright L. C., McClean P. E., et al. (2013). In silico comparison of genomic regions containing genes coding for enzymes and transcription factors for the phenylpropanoid pathway in Phaseolus vulgaris L. and Glycine max L. Merr. Front. Plant Sci. 4:317 10.3389/fpls.2013.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schmutz J., McClean P. E., Mamidi S., Wu G. A., Cannon S. B., Grimwood J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestication. Nat. Genet. 46, 707–713 10.1038/ng.3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sassaki N., Akemi O. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Yu O., Jez J. M. (2008). Nature's assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J. 54, 750–762 10.1111/j.1365-313X.2008.03436.x [DOI] [PubMed] [Google Scholar]