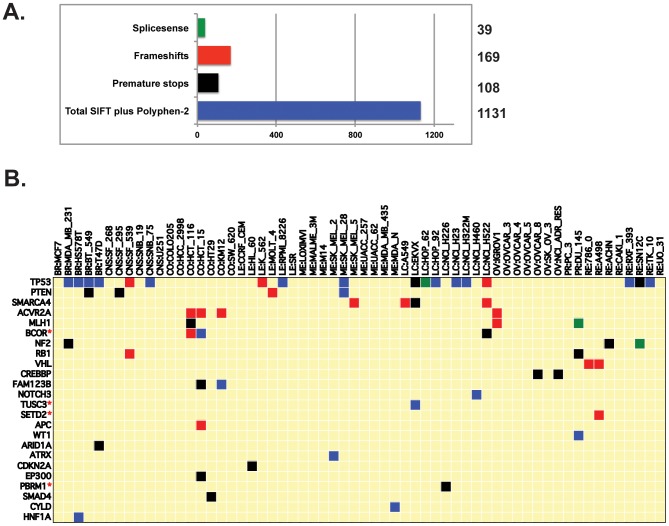
Figure 2. Homozygous, amino-acid changing, putative protein-function-affecting genetic variants present in the NCI-60, and absent in the 1000 Genomes and ESP5400.
A. The four categories of protein-function-affecting variants, and their level of occurrence. The x-axis is the number of variants in each category, with exact numbers given to the right. B. Potential knockout cell lines for tumor suppressors. The x-axis indicates the cell lines. The y-axis indicates the tumor suppressors. Green, red, black, and blue square indicate the presence of homozygous splicesense, frameshift, premature stop, and SIFT or PolyPhen-2 knockouts, respectively (as in A). Additional potential knockouts for the whole genome across the NCI-60 can be readily found in Table S1.