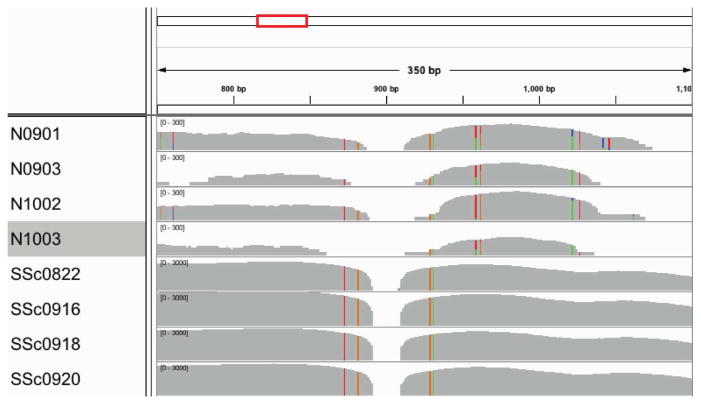
Figure 5. IGV visualization of original read set to R. glutinis 28S rRNA.
Aligning raw reads to R. glutinis 28S rRNA sequence (FJ345357) shows many reads aligning in the SSc samples but much fewer reads in the normal samples. The position shown is from 750–1100 in the sequence, which is the end of ITS2 and the first 400 bases of 28S rRNA. The gray histogram shows the depth of coverage at each base along the sequence; note that the axis is 10-fold higher for SSc samples. Colored bars show areas where the aligned reads differ from the reference sequence.