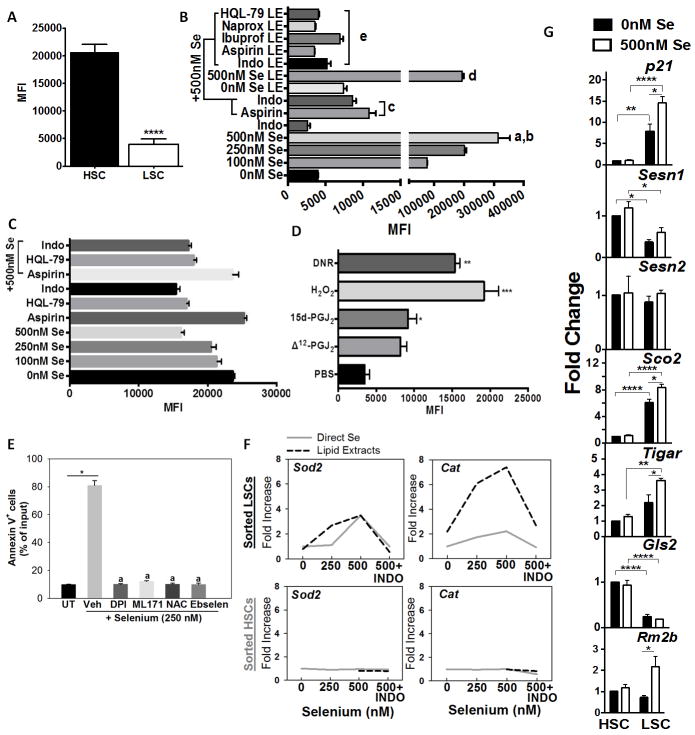
Figure 5.
Exacerbated production of ROS by supraphysiological selenium in BCR-ABL+LSCs. A, ROS levels in HSC and BCR-ABL+LSC prior to addition of selenium upon incubation with CellROX. Mean ± s.e.m of n= 3 independent assays; **** p<0.001. B, Changes in intracellular ROS in BCR-ABL+LSCs treated with various concentrations of exogenous selenium in the absence or presence of aspirin or indomethacin. LSCs were also treated with lipid extracts (LE) derived from the media supernatant of RAW264.7 macrophages cultured in the presence of various concentrations of selenium and HQL-79 or NSAIDs. All results shown are representative of n= 5–6 independent experiments per group. a, b represent P<0.001 compared to 0 nM Se and P <0.01 compared to 100 nM Se; c, P <0.01 compared to 500 nM Se; d, P <0.01 compared to 0 nM Se LE; e, P <0.05 compared to 500 nM Se LE. C, effect of addition of selenium to HSCs in the presence or absence of indomethacin, aspirin or HQL-79. Mean ± s.e.m of n=3 independent assays. D, Effects of Δ12-PGJ2 (25 nM) and 15d-PGJ2 (25 nM) on intracellular ROS in LSCs. LSCs were treated with CyPGs for 24 h; H2O2 (1 μM) for 30 min and DNR (1 μM) for 60 min. H2O2 and DNR were used as positive controls. E, effect of DPI (100 nM), ML171 (1μM, N-acetylcysteine (NAC; 1 mM), and ebselen (2 mM) on the apoptosis of sorted BCR-ABL+LSCs cultured in the presence of 250 nM selenium (as selenite) for 24 h. Cells were subjected to flow cytometric analysis with annexin V staining. *, a, p<0.01 compared to untreated (UT) or vehicle treated selenium group, respectively. F, dose-dependent effects of selenium in the presence or absence of indomethacin (INDO) on the expression of Sod2 and Cat in sorted HSCs and BCR-ABL+LSCs treated for 6 h. G, Selenium-dependent modulation of p21, Sesn1, Sesn2, Sco2, Tigar, and Rm2b in BCR-ABL+LSCs and HSCs. Cells were treated for 6 h following which mRNA was isolated and subjected to qPCR analysis. Data was normalized to 18 S rRNA and untreated HSCs were used to calculate fold changes. All data shown are mean ± s.e.m. of at least n=6 independent experiments. *, **, ***, **** represent p< 0.05, 0.01, 0.005, 0.001, respectively.