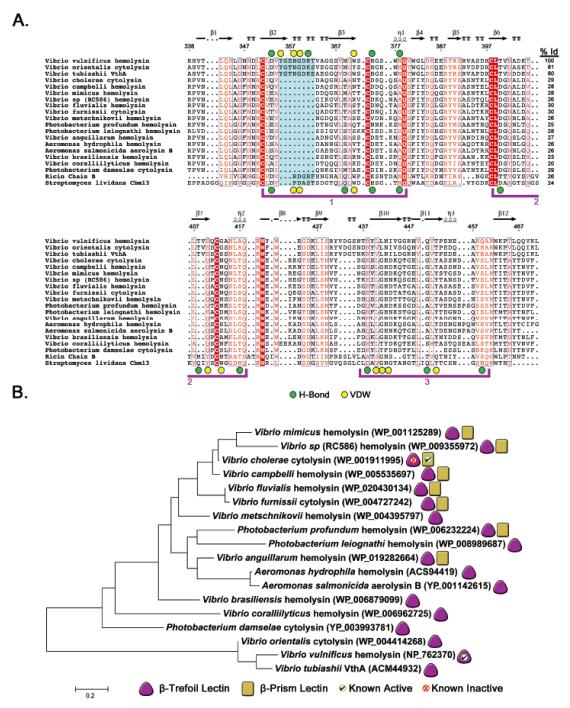
Figure 5. Sequence alignment of VVH and related β-trefoil domains.
A. A BLAST 36 search using the VVH β-trefoil protein sequence was conducted and representative sequences from related toxins aligned using the Muscle 52 algorithm as implemented in MEGA v. 5.2.2 38 and prepared using ESPript 3.0 (http://espript.ibcp.fr) 53. Sequences from β-trefoil domains from ricin B-chain and S. lividans CBM13 were included at the bottom of the alignment for reference. Three potential binding sites are bracketed in purple. The ricin domain has an active sugar-binding site in bracket 1 and CBM13 has three active binding sites. VVH residues involved in hydrogen bonding or van der Waals interactions are denoted as green or yellow circles, respectively, above the alignment. Residues involved in similar interactions in CBM13 are marked similarly below the alignment. The extended loop that contacts the penultimate carbohydrate unit in VVH is shaded in cyan and the degree of sequence identity between VVH and each toxin is listed in the right column. The numbers above the alignment are based on the complete VVH sequence and secondary structure is represented as calculated by ESPript. B. Evolutionary history as inferred using the maximum likelihood method based on the JTT matrix-based model as implemented in MEGA v. 5.2.2. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (111 total positions). Toxins with a β-trefoil domain are denoted with a purple triangle and the subset with β-prism domains are marked with a yellow rectangle. Domains for which we have obtained positive binding results using glycan-chip screening are marked with a green checkmark and domains that did not exhibit activity are marked with a red ‘X’.