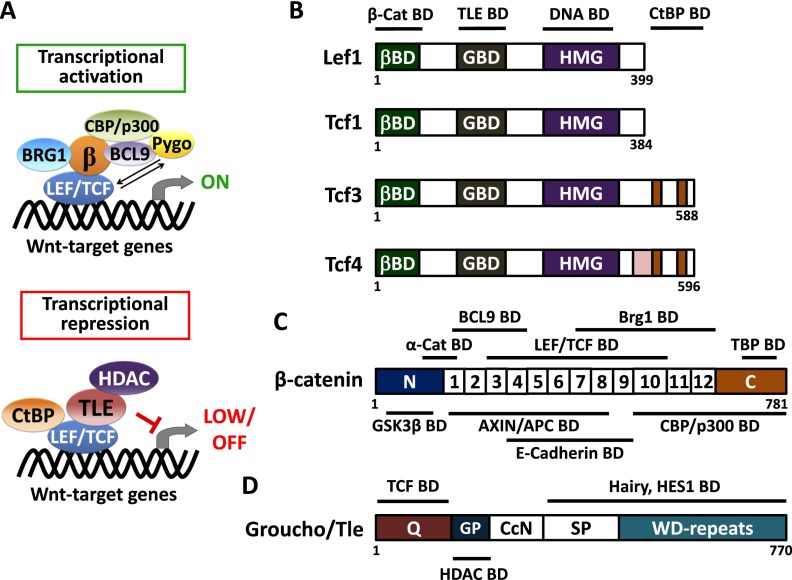
Figure 2.
Transcriptional regulation and structural organization of canonical Wnt regulators. (A) Schematic depicts a transcriptional activation or repression complex of LEF/TCF on Wnt target genes. In the activation mode, β-catenin interacts with a member of the LEF/TCF family of DNA-binding proteins. This conformation is thought to recruit histone modifiers CBP/p300 and BRG1 to yield an active chromatin structure for its target genes (Hecht et al. 2000; Takemaru and Moon 2000; Barker et al. 2001). Also participatory in this chromatin activation step is the H3K79 methyltransferase Dot1l, which, in the intestinal crypt, has been shown to be recruited to Wnt target genes in a β-catenin-dependent manner, thereby orchestrating broader chromatin remodeling and transcriptional elongation (Mahmoudi et al. 2010). Recruitment of BCL9 and Pygo are known to enhance β-catenin transactivator activity, although the mechanisms are still unfolding (Kramps et al. 2002; Thompson et al. 2002; Hoffmans and Basler 2004; Townsley et al. 2004; Li et al. 2007). Conversely, when nuclear β-catenin is absent, TCF3 and/or TCF4 proteins interact with transcriptional repressor Groucho/TLEs and in turn recruit histone deacetylase (HDAC) to yield an inactive chromatin state for the target genes. Another repressor, CtBP, has also been reported to interact with TCF4 for gene silencing (Cavallo et al. 1998; Brantjes et al. 2001; Valenta et al. 2003; Cuilliere-Dartigues et al. 2006; Arce et al. 2009; Cadigan and Waterman 2012). In general, whether LEF/TCF proteins act to activate or repress genes is determined by their binding partners and is cell context-dependent. (B–D) Structural organization of LEF/TCF member proteins, β-catenin, and Groucho/TLE protein. Line diagrams shown above and below the structures display binding domains for the indicated proteins. (B) LEF/TCF member proteins share a conserved structural organization, which consists of β-catenin-binding domain (βBD), putative Groucho/TLE-binding domain (GBD), and HMG DNA-binding domain. The CtBP-binding domain is seen for only TCF3 and TCF4 proteins. (C) The diagram displays the coactivator, β-catenin, which comprises 12 Armadillo repeats in the center of the protein structure. These repeats mediate most of the interactions between β-catenin and its binding partners, including the destruction complex components AXIN/APC, intercellular molecule E-cadherin, LEF/TCF transcription factors, histone modifier Brg1, and cofactor BCL9. The N-terminal region of β-catenin contains conserved phosphorylation residues for GSK3β for subsequent proteolytic degradation, and this region is also recognized by junctional protein α-catenin (Nelson and Nusse 2004). The C-terminal domain of β-catenin includes potent transcriptional transactivation elements that recruit coactivators TBP and CBP/p300. (D) The diagram depicts a model for the Groucho/TLE proteins (Chodaparambil et al. 2014). Most of them share a conserved structural organization, which consists of a glutamine-rich (Q) domain followed by a glycine/proline-rich (GP) domain, CcN domain, serine/proline-rich (SP) domain, and WD repeat domain. Recently, it was shown that TCF3 and TCF4 can bind to the N-terminal region of TLE1, while HDACs are thought to be recruited by TLEs through their GP domain. TCF1 and LEF1 appear to have weaker binding to TLEs, which may account for their more typical behavior as coactivators rather than repressors for Wnt signaling (Chodaparambil et al. 2014). Other than TCFs, Groucho/TLEs also interact with other transcriptional factors (e.g., HES1) through their C-terminal region (Grbavec et al. 1998).