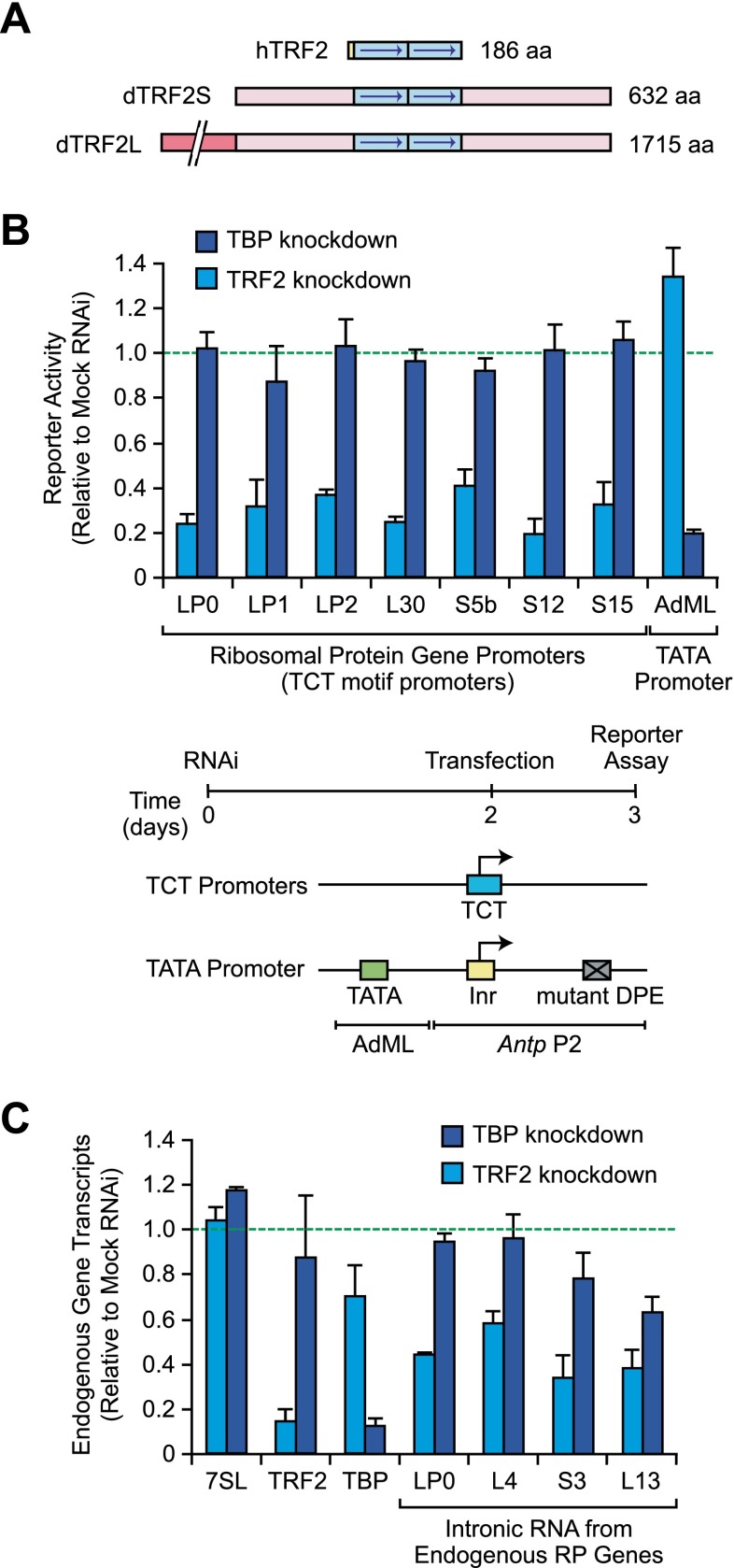
Figure 1.
TCT-dependent transcription appears to require TRF2 but not TBP. (A) Schematic diagrams of hTRF2 and the two forms of Drosophila TRF2 (dTRF2S and dTRF2L). dTRF2S is identical to the C-terminal 632-amino-acid residues of dTRF2L. (B) Depletion of TRF2, but not TBP, reduces RP gene expression. Drosophila S2 cells were depleted of either TRF2 or TBP by RNAi and then transfected with TCT-dependent or TATA-dependent luciferase reporter genes. The experimental scheme and reporter constructs are depicted at the bottom of the figure. The activities of the RNAi-depleted extracts are reported as relative to the activities of mock RNAi-treated control extracts. Error bars represent the standard deviation. (C) Analysis of endogenous transcript levels by qRT–PCR. Drosophila S2 cells were depleted of TRF2 or TBP, as in B. The total RNA was then isolated and analyzed by qRT–PCR. For the RP genes, intronic sequences were used to detect newly synthesized transcripts. We did not analyze RP genes with intronic snoRNA genes, as they could affect the levels of the intronic RNAs. The error bars represent the standard deviation.