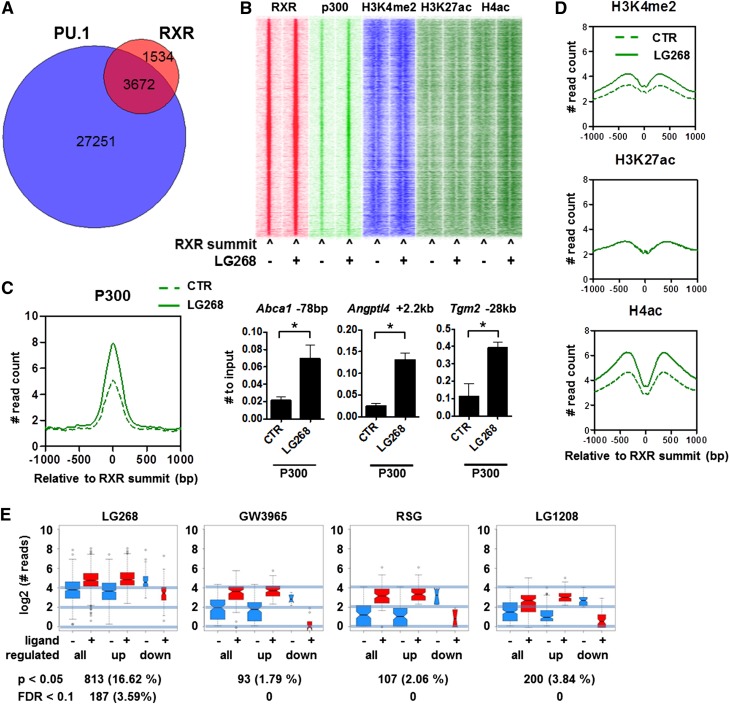
Figure 2.
The cistromic interactions of RXR, PU.1, P300, and active histone marks. (A) The intersection of cistromes as assessed by number of overlapping peaks is represented as a Venn diagram of RXR and PU.1. (B) Heat map representation of RXR, P300, H3K4me2, H3K27ac, and H4ac occupancies in 3-kb windows around the summit of the RXR peaks in the presence or absence of LG268. Read distribution was determined by HOMER, clustering was done by Gene Cluster 3.0 using centered correlation similarity metric with single linkage clustering method, and heat maps were created by Java TreeView in log2 scale. (C) Read distribution of P300 on RXR peaks in the presence or absence of LG268. P300 binding was confirmed on the indicated individual enhancers using ChIP-RT-qPCR. Macrophages were treated with 100 nM LG268 for 1 h. The mean and ±SD of three biological replicates are shown. Asterisk represents significant difference at P < 0.05; n = 3. (D) Average read distribution of the indicated active histone marks on RXR peaks in the presence or absence of LG268 determined by HOMER. (E) RXR enrichments of the significantly changing peaks in the presence of the indicated ligands. Cistrome was determined in the presence or absence of LG268, and the log2-normalized read numbers of the significantly changing peaks were plotted by DiffBind. The number of changing peaks and the statistical stringency applied is indicated below each plot.