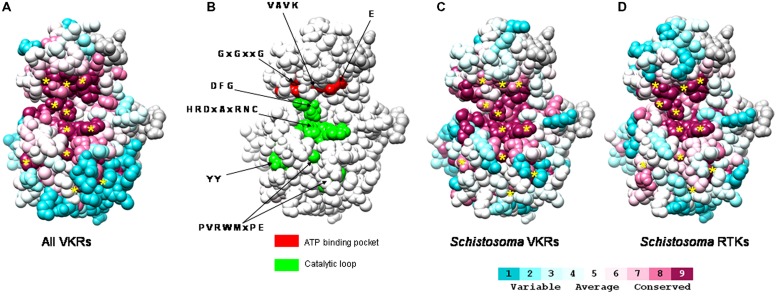
FIGURE 2.
Conservation of schistosome RTK tyrosine kinase catalytic sites. An evolutionary trace analysis of the conservation of residues in TK domains was performed using sequence alignment of (A) 40 VKRs already known in various invertebrate species (Vanderstraete et al., 2013a), (C) four VKRs of Schistosoma (SmVKR1 AAL67949.1 and SmVKR2 ADD91576.1, and two VKRs of Schistosoma haematobium Sha_103537 and Sha_104501), (D) various Schistosoma RTKs (EGF, insulin and VKR receptors) including SmVKR1, SmVKR2, the two ShVKRs, SmIR1 (AAN39120), SmIR2 (AAV65745.2), SjIR1 (ACT20714.1), SjIR2 (ACT20715.1), and SER (AAA29866.1). Visualization of the conservation was performed on the human IR TK crystal structure (PDB accession number 1IRK; Ahier et al., 2009). The alignment generated with ClustalW was submitted to the ConSurf website server (http://consurf.tau.ac.il; Landau et al., 2005). Conservation scores of each residue were calculated by taking into account the phylogenetic relationships among the sequences and the similarity between the amino acids in the alignment. Conservation scores are according to a color scale from variable (blue) to conserved (purple) residues. In (B), are indicated the crucial residues of the ATP binding pocket (red) and of the catalytic loop (green) required for kinase activity. The evolutionary trace analyses revealed that these crucial residues (indicated by * in A,C,D) are highly conserved among all VKRs (A,C) and among the panel of Schistosoma RTKs (EGF, insulin, and VKR receptors) from the three human species (mansoni, haematobium, and japonicum).