Figure 3. Identification of H3K4me3 and H3K27me3 Bivalent Chromatin Domains in QSCs.
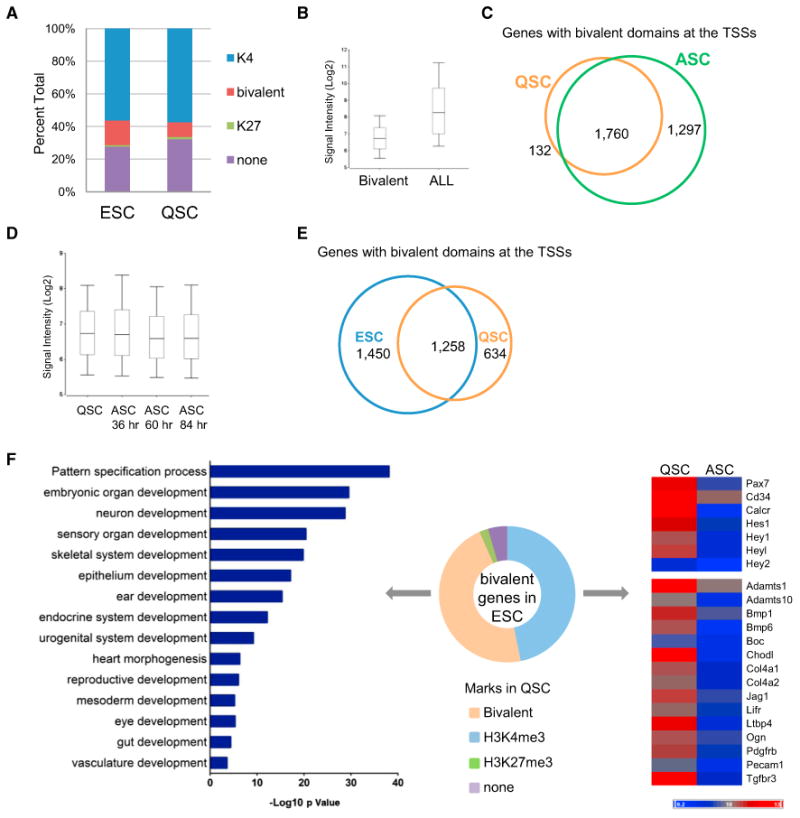
(A) Comparison of the proportion of genes marked by one of the four H3K4me3 and H3K27me3 patterns, H3K4me3 only (K4), H3K27me3 only (K27), both (bivalent), and neither (none), in ESCs and QSCs.
(B) Box and whisker plot of the expression level of genes that were found bivalent at the TSS in comparison to all genes in QSCs.
(C) Venn diagram of genes with bivalent domains at the TSS in QSCs and ASCs. Among the 1,892 genes marked by bivalent domains in QSCs, 1,760 were also found in ASCs.
(D) Box and whisker plot of the expression level of genes bivalent at the TSS in QSCs and ASCs of different time points in muscle regeneration.
(E) Venn diagram of genes with bivalent domains at TSSs in ESCs and QSCs.
(F) H3K4me3 and H3K27me3 patterns in QSCs of genes that are marked by bivalent domains in ESCs. The middle pie chart depicts the proportion of bivalent ESC genes that exhibited different H3K4me3 and H3K27me3 marks in QSCs. Among all genes that are bivalent in ESCs, 46% were found bivalent in QSCs (orange), and another 46% were found to be H3K4me3 only (blue). GO analysis of genes that were found bivalent in QSCs is shown by the bar graph on the left panel. The heatmap on the right panel depicts the expression level in QSCs and ASCs of representative genes that are bivalent at TSS in ESCs but H3K4me3 only in QSCs. The top panel of the heatmap shows genes known as QSC markers, and the bottom panel shows 15 of all 411 genes that encode glycoproteins (p = 10−39).
See also Figure S5.
