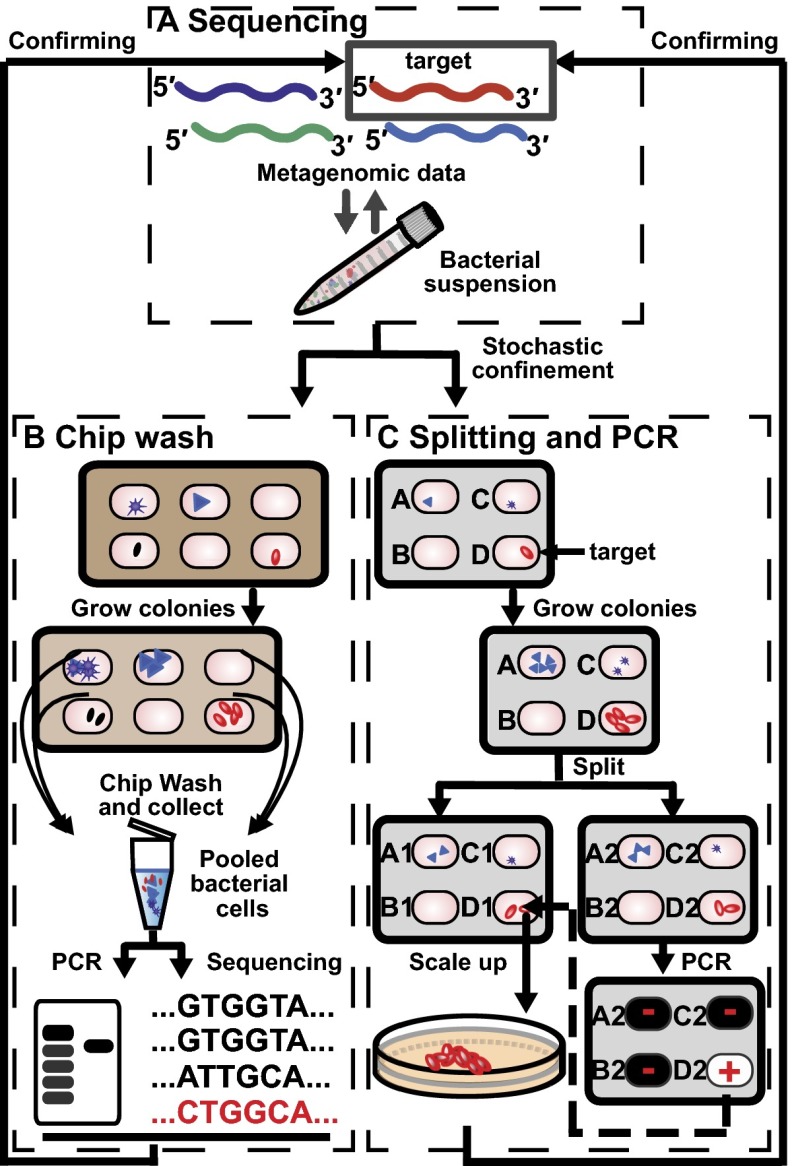
Fig. 1.
Illustration representing the workflow for targeted cultivation and isolation of microbial organisms. (A) Microbial targets carrying genes of interest are identified by high-throughput sequencing of clinical samples. A representative sequence of the target is shown in red. To cultivate the target, the inoculum is suspended in cultivation medium and loaded onto a microfluidic device, enabling stochastic confinement of single cells and cultivation of individual species (represented by different shapes). (B) A chip wash method is used to monitor bacterial growth under different cultivation conditions. Cells are pooled en masse into a tube and DNA is extracted for genetic analysis such as sequencing and PCR. (C) The target can be isolated by growing the sample under the growth condition identified from the chip wash. The two halves of the device are separated, resulting in two copies of each colony. On one half of the chip, target colonies are identified using PCR. Then, the target colony on the other half of the chip is retrieved for a scale-up culture, after which sequencing is used to validate that the correct target has been isolated.