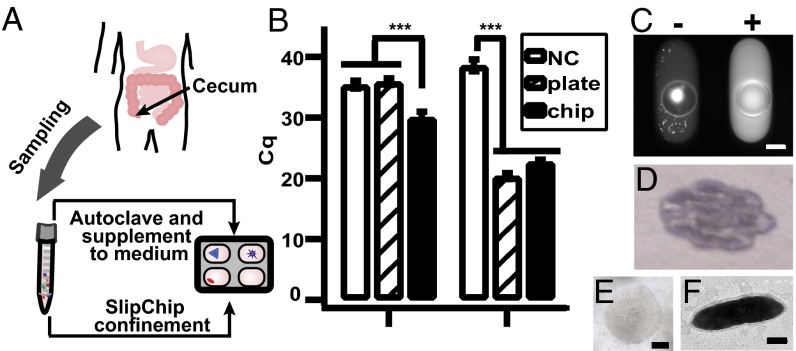
Fig. 5.
Targeted isolation of isolate microfluidicus 1 from SlipChip. (A) Illustration showing that mucosal biopsies obtained from the human cecum were used for stochastic confinement as well as supplemented into the medium to stimulate growth of microbes. (B) Identifying the cultivation condition of the microbial target OTU158 using qPCR. (Left) Graph showing that the use of target-specific primers revealed that the target was found in the chip wash solution (M2LC) but not in the blank negative control (NC) or the plate wash solution (M2GSC). (Right) Graph showing that the use of universal primers of 16S rRNA gene showed that both chip wash and plate wash solutions contained bacterial genomic DNA. A lower Cq value indicates higher concentration of DNA. Error bars indicate SD (n = 3). (C) Fluorescence microscopy photograph of on-chip colony PCR after the chip was split, showing a positive well (Right) for OTU158. A PCR negative well is shown on the left, as indicated by the low fluorescence intensity of the solution. The bright spot was presumably from cell material stained with SYBR Green. (D) Photograph of the first round of scaled-up culture of OTU158. (E) Microphotograph of a single colony of isolate microfluidicus 1. (F) Transmission electron microscopy image of a single OTU158 cell. Scale bar, 200 µm for C and E, and 0.5 µm for F.